豬miR-124靶向IQGAP2調節巨噬細胞內沙門氏菌的增殖
陳旺 鄧榆 殷俊 官州 金凱 石博妹 黃廷華 姚敏
摘要:【目的】明確豬miR-124與其靶基因IQGAP2間的表達調控關系,以及miR-124表達水平與豬巨噬細胞內沙門氏菌數量的關聯,為揭示沙門氏菌在感染細胞內存活與增殖的機制提供理論依據。【方法】通過熒光素酶報告基因系統驗證miR-124與IQGAP2基因的作用位點;再以GM-CSF誘導的豬巨噬細胞和鼠傷寒沙門氏菌(ATCC 14028)為試驗材料,通過實時熒光定量PCR和流式細胞術測定沙門氏菌感染豬巨噬細胞中miR-124和IQGAP2基因的表達及巨噬細胞內沙門氏菌的增殖情況。【結果】miR-124結合位點野生型載體轉染的熒光報告信號顯著低于miR-124結合位點突變載體(P<0.05,下同),但共轉染anti-miR-124序列后能顯著增強miR-124結合位點野生型載體的熒光報告信號。經沙門氏菌感染后,豬巨噬細胞中的miR-124表達被激活,感染12、24和48 h后的相對表達量均顯著高于沙門氏菌感染前(0 h),而IQGAP2基因表達水平呈顯著下調趨勢;在沙門氏菌感染豬巨噬細胞內,miR-124表達水平與IQGAP2基因表達水平呈明顯負相關(r=-0.92)。miR-124高表達組細胞內的沙門氏菌數量顯著高于正常巨噬細胞,但miR-124敲低表達組細胞內的沙門氏菌數量顯著低于正常巨噬細胞;IQGAP2基因敲低表達組細胞內的沙門氏菌數量顯著高于正常巨噬細胞;此外,miR-124高表達+IQGAP2基因敲低表達組細胞內的沙門氏菌數量與IQGAP2基因敲低表達處理組相比無顯著差異(P>0.05),但顯著高于miR-124高表達組細胞。【結論】沙門氏菌感染豬巨噬細胞中的miR-124表達水平與IQGAP2基因表達水平及胞內沙門氏菌數量呈負相關,即沙門氏菌可通過上調miR-124表達靶向抑制IQGAP2基因表達,從而調節其在豬巨噬細胞內的增殖。
關鍵詞: 豬沙門氏菌;miR-124;IQGAP2基因;胞內增殖;流式細胞術;熒光素酶報告基因系統
中圖分類號: S852.61? ? ? ? ? ? ? ? ? ? ? ? ? ? ? 文獻標志碼: A 文章編號:2095-1191(2020)12-3066-07
Abstract:【Objective】To explore the regulation relationship between miR-124 and its target gene IQGAP2, study the relationship between the expression level of miR-124 and the number of Salmonella in pig macrophages,so as to provide a theoretical basis for revealing the mechanism of survival and proliferation of Salmonella in infected cells. 【Method】The binding sitebetween miR-124 and IQGAP2 was verified by luciferase reporter gene system assay. Then, the GM-CSF induced macrophages and Salmonella typhimurium(ATCC 14028) were used as research materials. The expression of miR-124 and IQGAP2 gene and the proliferation of Salmonella in macrophages were determined by real-time quantitative PCR and flow cytometry. 【Result】The fluorescence signal of wild-type vector transfected by miR-124 binding site was significantly lower than that of miR-124 binding site mutant vector(P<0.05, the same below), and the co-transfection of anti-miR-124 sequence could significantly enhance the fluorescence signal of wild-type vector. In the process of Salmonella infection, the expression of miR-124 was activated,the relative expression levels at 12, 24 and 48 h after infection were significantly higher than those before infection (0 h). The expression of IQGAP2 was significantly down-regulated. In Salmonella infected macrophages, the expression levels of miR-124 were negatively correlated with IQGAP2 (r=-0.92). The Salmonella counts in miR-124 high expression group were significantly higher than the control. The Salmonella counts in miR-124 knock down group were significantly lower than the control macrophages. The Salmonella counts in miR-124 knock-down group were significantly higher than the control macrophagesgroup. The numberof Salmonella counts in miR-124 high expression+IQGAP2 gene knockdown expression group were not significantly different compared to the IQGAP2 knockdown group(P>0.05), but significantly higher than the miR-124 high expression group cells. 【Conclusion】The expression levels of miR-124 and IQGAP2 in Salmonella infected porcine macrophages are negatively correlated with intercellular Salmonella counts. Salmonella inhibits IQGAP2 gene expression by up-regulating miR-124 expression targeting, thereby regulating its proliferation in porcine macrophages.
Key words: porcine Salmonella; miR-124; IQGAP2 gene; intracellular proliferation; flow cytometry; luciferase reporter gene system
Foundation item: National Natural Science Foundation of China(31902231,31402055); Innovation and Entrepreneurship Program of Yangtze University (2018057)
0 引言
【研究意義】沙門氏菌(Salmonella)感染豬群后通常形成急性一過性反應和長期攜帶2種狀態(Alban and St?rk,2005;Bonardi,2017),即攜帶沙門氏菌生豬是沙門氏菌傳播感染的重要媒介(Guan and Holley,2003;李帆等,2018)。沙門氏菌是一種常見的人畜共患病原菌(施開創等,2018),在我國的動物源性食物中毒事件中有70%~80%是由沙門氏菌污染豬肉產品而引起(楊懷珍等,2016);沙門氏菌在感染細胞(主要為吞噬細胞)內存活并實現胞內增殖是其形成長期攜帶及傳播致病的重要原因(Lathrop et al.,2015)。因此,研究沙門氏菌在感染細胞內的存活和增殖發生機理及揭示相關基因在該過程中的調控作用,可為豬沙門氏菌感染的抗病育種提供分子標記,對防控生豬養殖生產中的沙門氏菌感染具有重要意義。【前人研究進展】本課題組前期通過高通量測序分析發現,在沙門氏菌感染仔豬外周血中miR-124、miR-16、miR-155和miR-143等29個miRNA呈顯著差異表達,其中miR-124在沙門氏菌感染仔豬外周血中呈顯著上調表達(5.86倍)(Huang et al.,2019),暗示miR-124是沙門氏菌感染過程中的重要調節因子。miR-124在免疫器官和免疫細胞,如外周血單核細胞、骨髓、淋巴結和胸腺組織中高表達,故推測其廣泛參與機體的免疫調節過程(Smerkova et al.,2015)。miR-124可被脂多糖(LPS)和牛分枝桿菌卡介苗誘導,其轉錄水平受TLR信號適配分子MyD88調控(Mehta and Baltimore,2016;Sun et al.,2016)。激活后的miR-124通過阻礙TLR6、MyD88、TNF-α、USP2、USP14、P65、TRAF6及STAT3而反作用于(抑制)TLR信號通路(Sun et al.,2013;Ma et al.,2014;Qiu et al.,2015),其在基因組中存在多個拷貝,分別為miR-124-1、miR-124-2和miR-124-3,雖然三者的前體序列有所不同,但其成熟序列在人類、小鼠和豬中完全一致,說明miR-124在不同物種體內發揮著相同的生物學功能。沙門氏菌感染仔豬外周血miRNA/mRNA表達譜數據聯合分析發現,在miR-124作用的110個候選靶基因中,沙門氏菌感染信號通路相關基因IQGAP2下調5倍,但同家族的IQGAP1和IQGAP3基因均未見顯著差異表達(Huang et al.,2019)。IQGAP2基因通過與CDC42-GTP相結合并調節CDC42的活化狀態(Brill et al.,1996),活化的CDC42進一步激活MAPK信號通路(Chen et al.,1996;Hobbie et al.,1997;Patel and Galán,2006),進而調節宿主的免疫功能(Garrett et al.,2000;Rodriguez-Escudero et al.,2011;Lathrop et al.,2015)。【本研究切入點】miR-124是沙門氏菌感染過程中的重要調節因子,是沙門氏菌在宿主體內存活、建立攜帶狀態及感染致病的關鍵因素,但至今有關miR-124對下游靶基因IQGAP2的調節作用及其對細胞內沙門氏菌增殖情況的影響機制尚未明確。【擬解決的關鍵問題】采用實時熒光定量PCR、流式細胞術及熒光素酶報告檢測系統對沙門氏菌感染豬巨噬細胞中miR-124和IQGAP2基因的表達及巨噬細胞內沙門氏菌的增殖情況進行探討,為揭示沙門氏菌在感染細胞內的存活與增殖機制提供理論依據。
1 材料與方法
1. 1 試驗材料
胎牛血清(10100154)和細胞培養液RPMI 1640(22400105)購自美國Gibco公司;哺乳動物外周血單核細胞分離液Percoll(P1644)購自Sigma-Aldrich公司;重組豬GM-CSF(ab233683)購自英國Abcam公司;總RNA提取試劑盒(R1200)購自北京索萊寶科技有限公司;miR-124定量檢測試劑盒(480901_mir)購自美國ABI公司;兔抗人IQGAP2多克隆抗體(PA5-95484)和PE標記的羊抗兔IgG(31864)購自ThermoFisher公司;FITC標記的羊抗人CD14抗體(ABIN2478467)購自北京四正柏生物科技有限公司;動物細胞轉染試劑TransFastTM(E2431)購自美國Promega公司;IQGAP2基因siRNA序列、miR-124序列、anti-miR-124序列及實時熒光定量PCR擴增引物(表1)均委托生工生物工程(上海)股份有限公司合成;psiCHECKTM-1、pGL3-Control、鼠傷寒沙門氏菌(ATCC 14028)和Raw264.7細胞由長江大學動物科學學院分子生物學實驗室保存提供。
1. 2 miR-124高表達或敲低表達巨噬細胞的建立
以無菌生理鹽水對仔豬外周抗凝血進行1∶1稀釋,再使用哺乳動物外周血單核細胞分離液Percoll分離外周血單核細胞,并轉移至細胞6孔培養板中,經貼壁、洗滌后,以GM-CSF(20 ng/mL)和IL-4(10 ng/mL)刺激培養6 d,以獲得充分分化的巨噬細胞。將IQGAP2 siRNA(10 nmol/L)、miR-124 mimics(10 nmol/L)和anti-miR-124(10 nmol/L)以TransFastTM轉染試劑包裹后分別轉染分化的巨噬細胞,傳染24 h后使用實時熒光定量PCR檢測細胞中IQGAP2基因和miR-124的轉錄水平,以證實各組細胞建模是否成功。將對數生長期的沙門氏菌與構建的各組模型細胞按1∶1比例互作侵染,分別于感染0、12、24和48 h時收集細胞樣品用于提取總RNA,設3次重復。采用TRIzol法提取總RNA,利用Stem-Loop TaqMan檢測總RNA中的miR-124表達情況,以SYBR-Green PCR Master Mix試劑盒檢測總RNA中的IQGAP2基因表達水平。用4%甲醛重懸細胞并在室溫(20~25 ℃)下固定15 min,1×PBS離心洗滌。向預冷的細胞中緩慢加入預冷的100%甲醇,至甲醇終濃度為90%,溫和渦旋以通透細胞。BD-PharmingenTM染色緩沖液以抗CD14-FITC抗體進行表面表型染色30 min,然后將通透處理的細胞重懸于BD PharmingenTM染色緩沖液中,加入anti-IQGAP2抗體,室溫下孵育20 min。以100 ?L稀釋的羊抗兔IgG偶聯PE二抗再次重懸細胞,并室溫孵育30 min,最后通過BD-LSRFortessaTM流式細胞分析儀進行分析。每組細胞處理均設3次重復,流式細胞術數據提交至流庫數據庫(Spidlen et al.,2012),編號FR-FCM-Z2FQ。
1. 3 胞內沙門氏菌增殖模型
參考Huang等(2018,2019)的方法以沙門氏菌侵染巨噬細胞。正常巨噬細胞、miR-124高表達及敲低表達巨噬細胞、IQGAP2基因敲低表達巨噬細胞、miR-124高表達+IQGAP2基因敲低表達巨噬細胞及miR-124敲低表達+IQGAP2基因敲低表達巨噬細胞經PBS洗滌和重懸后,分別按1×106/孔的密度接種至細胞6孔培養板中,同時按1∶1比例將對數生長期的沙門氏菌接種至各組巨噬細胞中,細胞培養板500 r/min離心10 min,使細胞和細菌聚集于培養底面,在37 ℃、5% CO2培養箱中孵育2 h,PBS漂洗3次,加入Gentamicin培養基(100 ?g/mL)繼續培養2 h,再使用無抗生素培養基繼續培養一定時間后收集細胞樣品。自沙門氏菌與巨噬細胞互作時開始計時,分別收集4、8和12 h時的細胞樣品,各3份。采用1% Tritone裂解破碎巨噬細胞,抽提DNA,使用實時熒光定量PCR檢測沙門氏菌數量(Vinayaka et al.,2019)。
1. 4 雙螢光素酶報告分析
將含有miR-124結合位點(5189~5195 nt,XM_021084462)的IQGAP2基因3'-UTR序列(XM_0210 84462,4761~5860 nt)野生型或突變(5192C/G)片段克隆至psiCHECKTM-1載體質粒海腎螢光素酶ORF下游,分別命名為psiCHECKTM-1-IQGAP2 3'-UTR(Wild-type)和psiCHECKTM-1-IQGAP2 3'-UTR(Mutant)。將RAW264.7細胞以每孔0.5×106個細胞接種至細胞6孔培養板中,當有60%細胞融合時采用TransFastTM轉染試劑包裹miR-124(或anti-miR-124)、psiCHECKTM-1-IQGAP2 3'-UTR(Wild-type)[或psiCHECKTM-1-IQGAP2 3'-UTR(Mutant)],轉染Raw264.7細胞。試驗共設4組:(1)Mutant;(2)Mutant+miR-124;(3)Wilde type;(4)Wilde type+anti-miR-124。所有細胞均與轉染效率對照質粒pGL3-Control共轉染。每轉染4 h,即采用雙螢光素酶報告分析系統測量螢光素酶活性1次(Huang et al.,2018)。每組重復3次。
1. 5 統計分析
試驗數據采用SPSS 18.0進行統計分析,其中,兩組間比較采用 t 檢驗,多組間比較采用單因素方差分析(One-way ANOVA)。
2 結果與分析
2. 1 雙螢光素酶報告解析miR-124對IQGAP2基因的調控作用
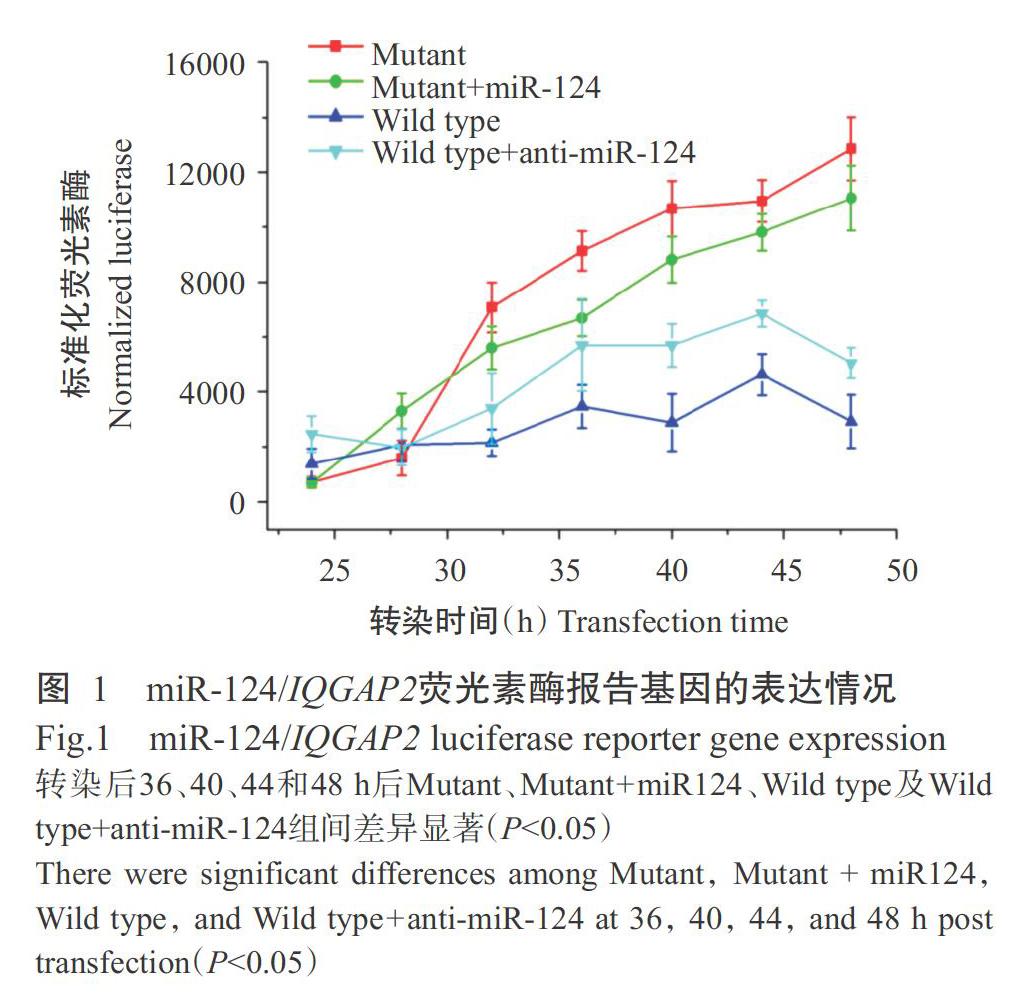
將IQGAP2基因3'-UTR區插入熒光素酶基因3'-UTR區,構建靶基因3'-UTR區的熒光素酶報告基因載體,以構建好的熒光素酶報告基因載體轉染Raw264.7細胞,通過檢測熒光素酶的表達情況分析IQGAP2基因3'-UTR區是否含有miR-124靶位點;同時構建miR-124結合位點人工點突變的報告基因載體。通過比較野生型和突變型報告基因的熒光素酶活性(圖1)發現,轉染36~48 h后miR-124結合位點野生型載體轉染的熒光報告信號顯著低于miR-124結合位點突變載體(P<0.05,下同),但共轉染anti-miR-124序列后能顯著增強miR-124結合位點野生型載體的熒光報告信號;miR-124結合位點突變載體轉染及anti-miR-124序列與miR-124結合位點突變載體共轉染均顯著增強報告基因表達,說明miR-124能作用于IQGAP2基因3'-UTR區而抑制報告基因表達。
2. 2 沙門氏菌感染豬巨噬細胞中miR-124和IQGAP2基因的轉錄水平
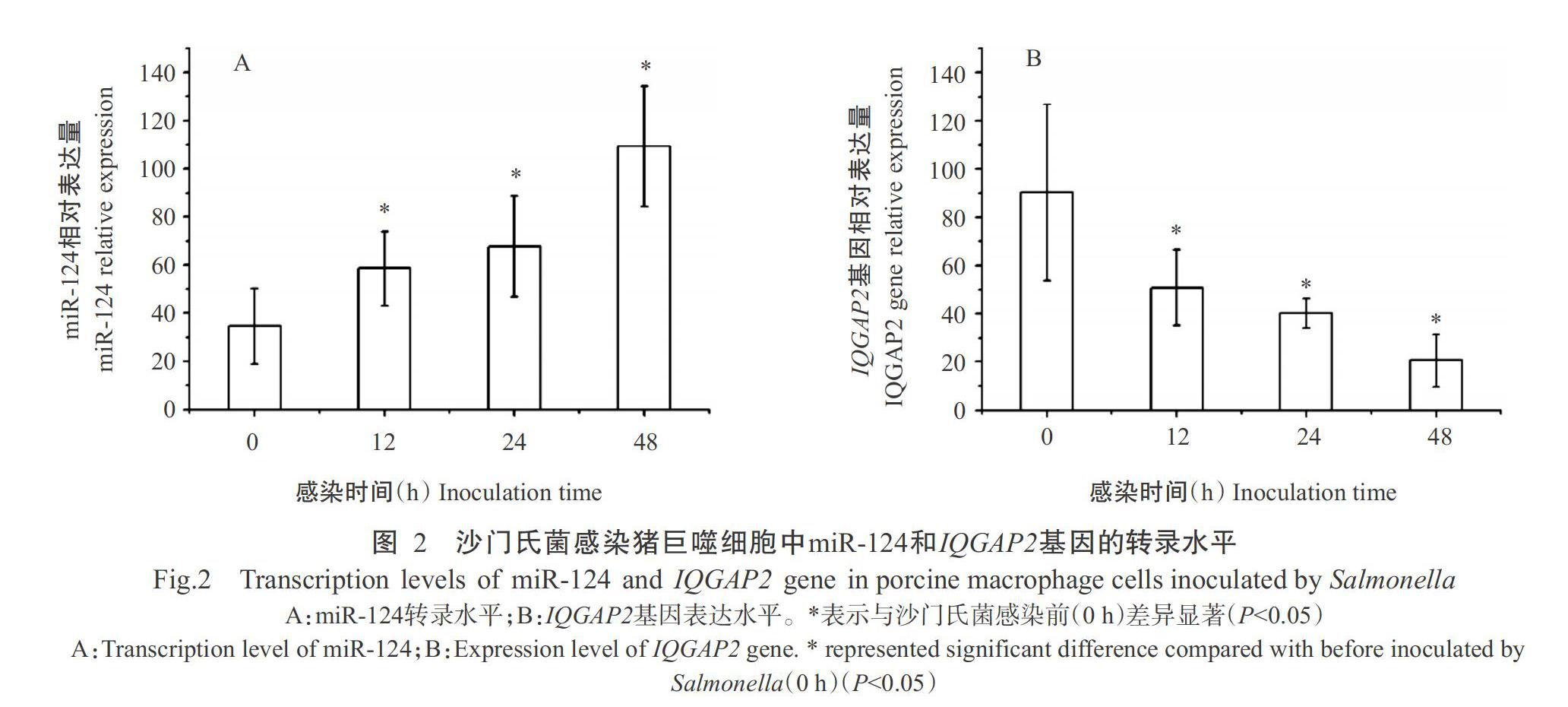
經沙門氏菌感染后,豬巨噬細胞中的miR-124表達水平明顯升高(圖2-A),感染12、24和48 h后的相對表達量均顯著高于沙門氏菌感染前(0 h);IQGAP2基因表達水平則明顯下調(圖2-B),感染12、24和48 h的相對表達量顯著低于沙門氏菌感染前(0 h)。在沙門氏菌感染豬巨噬細胞內,miR-124表達水平與IQGAP2基因表達水平呈明顯負相關(r=-0.92)。
2. 3 沙門氏菌感染豬巨噬細胞中IQGAP2蛋白的流式細胞術檢測結果
經沙門氏菌感染后,豬巨噬細胞中的IQGAP2蛋白表達水平呈明顯下調趨勢。在沙門氏菌感染前(0 h)約有61.1%的IQGAP2陽性細胞,沙門氏菌感染12 h后約有43.6%的IQGAP2陽性細胞,感染24 h后約有38.2%的IQGAP2陽性細胞,感染48 h后僅有23.0%的IQGAP2陽性細胞。即沙門氏菌感染12和24 h后的IQGAP2陽性細胞約是沙門氏菌感染前(0 h)的2/3,感染48 h后的IQGAP2陽性細胞約為沙門氏菌感染前(0 h)的1/3。流式細胞術檢測結果見圖3。
2. 4 沙門氏菌感染豬巨噬細胞內沙門氏菌數量的增殖規律
在沙門氏菌感染4 h后, miR-124高表達組細胞內的沙門氏菌數量明顯高于正常巨噬細胞(約1.5倍),IQGAP2基因敲低表達組細胞內的沙門氏菌數量則是正常巨噬細胞的3.5倍。至沙門氏菌感染8 h后,miR-124高表達組和IQGAP2基因敲低表達組細胞內的沙門氏菌數量持續上升,miR-124高表達組細胞內的沙門氏菌數量明顯高于正常巨噬細胞(約1.8倍),IQGAP2基因敲低表達組細胞內的沙門氏菌數量是正常巨噬細胞的4.0倍。在沙門氏菌感染12 h后,各處理組的胞內沙門氏菌數量均較感染8 h時有所上升,miR-124高表達組細胞內的沙門氏菌數量明顯高于正常巨噬細胞(約2.0倍),IQGAP2基因敲低表達組細胞內的沙門氏菌數量是正常巨噬細胞的4.5倍。各感染時間點,miR-124敲低表達組胞內沙門氏菌數量均顯著低于正常巨噬細胞;miR-124高表達+IQGAP2基因敲低表達組細胞內的沙門氏菌數量與IQGAP2基因敲低表達處理組相比無顯著差異(P>0.05,下同),但顯著高于miR-124高表達組細胞;miR-124敲低表達+IQGAP2基因敲低表達組細胞內的沙門氏菌數量與IQGAP2基因敲低表達處理組相比也無顯著差異,但顯著高于miR-124敲低表達組細胞、低于miR-124高表達+IQGAP2基因敲低表達組細胞。
3 討論
沙門氏菌通過糞口途徑進入動物體內,部分酸耐受菌體從胃進入小腸,穿過小腸黏膜層而作用于黏膜下層的M細胞(Prouty et al.,2004),再轉運至腸系膜淋巴結(Chen et al.,2015)。沙門氏菌進入動物體內后,主要通過以下3條途徑與機體免疫系統相互作用:(1)來源于沙門氏菌的LPS在MD2和CD14輔助下能被TLR4受體識別,TLR4識別沙門氏菌后可激活巨噬細胞并作用于TLR4適配分子MyD88,從而激活下游TLR4信號通路和NFκB信號通路(Mastroeni et al.,2009;Deng et al.,2016);(2)沙門氏菌通過III型分泌系統將菌體蛋白分泌到動物細胞內,直接(SopE和SopE2)或間接(SopB)作用于CDC42,并進一步激活MAPK信號通路(Chen et al.,1996;Hobbie et al.,1997;Patel and Galán,2006);(3)沙門氏菌基因組DNA可被動物細胞的外源DNA識別蛋白(DAI、AIM2和RIG-I)所識別,進而激活細胞質DNA感知信號通路(Qiu et al.,2015;Mehta and Baltimore,2016)。已有的研究雖然從不同角度闡明沙門氏菌對動物機體免疫系統的激活機制,但仍然無法合理解釋沙門氏菌免疫逃避甚至攜帶排菌的現象。
miR-124在動物免疫器官和免疫細胞,如外周血單核細胞、骨髓、淋巴結和胸腺中高表達,并廣泛參與機體免疫調節過程(Smerkova et al.,2015)。本研究結果表明,CSF誘導豬巨噬細胞經沙門氏菌感染后,miR-124呈上調表達,其上調表達機制可能與前人的相關研究結論(Mehta and Baltimore,2016;Sun et al.,2016)一致,即受沙門氏菌LPS-TLR信號的激活。IQGAP2基因的表達受甲基化和轉錄后水平等多種因素影響(Jin et al.,2008;Deng et al.,2016;Pelossof et al.,2016)。本研究的熒光素酶報告分析系統檢測結果顯示,IQGAP2基因3'-UTR區存在miR-124作用靶位點,一定程度上揭示了沙門氏菌感染仔豬巨噬細胞中miR-124與IQGAP2基因表達的負調控關系。miR-124高表達及IQGAP2基因敲低表達組豬巨噬細胞內沙門氏菌數量顯著高于miR-124敲低表達組細胞,暗示miR-124可影響豬巨噬細胞內的沙門氏菌數量,其機制可能是通過作用于IQGAP2信號而影響宿主細胞對抗原的吞噬和加工能力,從而更有利于胞內沙門氏菌的存活與增殖。因此,miR-124/IQGAP2路徑可能是沙門氏菌在宿主體內存活、增殖及建立攜帶狀態的重要因素,是沙門氏菌激活宿主免疫的剎車系統,但具體作用機制尚需進一步研究證實。
4 結論
沙門氏菌感染豬巨噬細胞中的miR-124表達水平與IQGAP2基因表達水平及胞內沙門氏菌數量呈負相關,即沙門氏菌可通過上調miR-124表達靶向抑制IQGAP2基因表達,從而調節其在豬巨噬細胞內的增殖。
參考文獻:
李帆,羅行煒,劉建華,梁軍,賀丹丹,潘玉善,苑麗,胡功政. 2018. 豬源沙門氏菌多藥外排泵oqxAB和氟苯尼考耐藥基因floR的檢測分析[J]. 江西農業學報,30(11):82-85. [Li F,Luo X W,Liu J H,Liang J,He D D,Pan Y S,Yuan L,Hu G Z. 2018. Detection and analysis of multi-drug efflux pump oqxAB and florfenicol-tolerant gene floR in swine-derived Salmonella typhimurium[J]. Acta Agriculturae Jiangxi,30(11):82-85.]
施開創,尹彥文,溫麗霞,屈素潔,王海清,胡杰. 2018. 黏菌素耐藥基因mcr-1 TaqMan-MGB熒光定量PCR檢測方法的建立[J]. 南方農業學報,49(7):1447-1452. [Shi K C,Yin Y W,Wen L X,Qu S J,Wang H Q,Hu J. 2018. Establishment of TaqMan-MGB fluorescent quantitative PCR for detection of colistin resistance gene mcr-1[J]. Journal of Southern Agriculture,49(7):1447-1452.]
楊懷珍,牟亞,羅薇. 2016. 食源性沙門氏菌的研究進展[J]. 黑龍江畜牧獸醫,(7):69-71. [Yang H Z,Mou Y,Luo W. 2016. Research progress of food-borne Salmonella[J]. Heilongjiang Animal Science and Veterinary Medicine,(7):69-71.]
Alban L,St?rk K D C. 2005. Where should the effort be put to reduce the Salmonella prevalence in the slaughtered swine carcass effectively?[J]. Preventive Veterinary Medi-cine,68(1):63-79.
Bonardi S. 2017. Salmonella in the pork production chain and its impact on human health in the European Union[J]. Epidemiology and Infection,145(8):1513-1526.
Brill S,Li S,Lyman C W,Church D M,Wasmuth J J,Weissbach L,Bernards A,Snijders A J. 1996. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmo-dulin and Rho family GTPases[J]. Molecular and Cellular Biology,16(9):4869-4878.
Chen J,Tian J,Tang X Y,Rui K,Ma J,Mao C M,Liu Y Z,Lu L W,Xu H X,Wang S J. 2015. miR-346 regulates CD4+CXCR5+ T cells in the pathogenesis of Graves? disease[J]. Endocrine,49(3):752-760.
Chen L M,Hobbie S,Galán J E. 1996. Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses[J]. Science,274(5295):2115-2118.
Deng Z,Wang L J,Hou H L,Zhou J C,Li X. 2016. Epigenetic regulation of IQGAP2 promotes ovarian cancer progression via activating Wnt/beta-catenin signaling[J]. International Journal of Oncology,48(1):153-160.
Garrett W S,Chen L M,Kroschewski R,Ebersold M,Turley S,Trombetta S,Galán J E,Mellman I. 2000. Developmental control of endocytosis in dendritic cells by Cdc42[J]. Cell,102(3):325-334.
Guan T Y,Holley R A. 2003. Pathogen survival in swine manure environments and transmission of human enteric illness—A review[J]. Journal of Environmental Quality,32(2):383-392.
Hobbie S,Chen L M,Davis R J,Galán J E. 1997. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells[J]. Journal of Immunology,159(11):5550-5559.
Huang T H,Huang X L,Chen W,Yin J,Shi B M,Wang F F,Feng W Z,Yao M. 2019. microRNA responses associa-ted with Salmonella enterica serovar typhimurium challenge in peripheral blood:Effects of miR-146a and IFN-gamma in regulation of fecal bacteria shedding counts in pig[J]. BMC Veterinary Research,15(1):195. doi:10.1186/ s12917-019-1951-4.
Huang T H,Huang X L,Yao M. 2018. miR-143 inhibits intracellular Salmonella growth by targeting ATP6V1A in macrophage cells in pig[J]. Research in Veterinary Science,117:138-143.
Jin S H,Akiyama Y,Fukamachi H,Yanagihara K,Akashi T,Yuasa Y. 2008. IQGAP2 inactivation through aberrant promoter methylation and promotion of invasion in gastric cancer cells[J]. International Journal of Cancer,122(5):1040-1046.
Lathrop S K,Binder K A,Starr T,Cooper K G,Chong A,Carmody A B,Steele-Mortimer O. 2015. Replication of Salmonella enterica serovar Typhimurium in human monocyte-derived macrophages[J]. Infection and Immunity,83(7):2661-2671.
Ma C Y,Li Y,Li M,Deng G C,Wu X L,Zeng J,Hao X J,Wang X P,Liu J,Cho W C S,Liu X M,Wang Y J. 2014. microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection[J]. Molecular Immunology,62(1):150-158.
Mastroeni P,Grant A,Restif O,Maskell D. 2009. A dynamic view of the spread and intracellular distribution of Salmonella enterica[J]. Nature Reviews. Immunology,7(1):73-80.
Mehta A,Baltimore D. 2016. microRNAs as regulatory elements in immune system logic[J]. Nature Reviews. Immunology,16(5):279-294.
Patel J C,Galán J E. 2006. Differential activation and function of Rho GTPases during Salmonella-host cell interactions[J]. The Journal of Cell Biology,175(3):453-463.
Pelossof R,Chow O S,Fairchild L,Smith J J,Setty M,Chen C T,Chen Z B,Egawa F,Avila K,Leslie C S,Garcia-Aguilar J. 2016. Integrated genomic profiling identifies microRNA-92a regulation of IQGAP2 in locally advanced rectal cancer[J]. Genes,Chromosomes & Cancer,55(4):311-321.
Prouty A M,Brodsky I E,Falkow S,Gunn J S. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium[J]. Microbiology(Rea-ding),150(Pt 4):775-783.
Qiu S W,Feng Y M,LeSage G,Zhang Y,Stuart C,He L,Li Y,Caudle Y,Peng Y,Yin D L. 2015. Chronic morphine-induced microRNA-124 promotes microglial immunosuppression by modulating P65 and TRAF6[J]. Journal of Immunology,194(3):1021-1030.
Rodriguez-Escudero I,Ferrer N L,Rotger R,Cid V J,Molina M. 2011. Interaction of the Salmonella typhimurium effector protein SopB with host cell Cdc42 is involved in intracellular replication[J]. Molecular Microbiology,80(5):1220-1240.
Smerkova K,Hudcova K,Vlahova V,Vaculovicova M,Peka-rik V,Masarik M,Adam V,Kizek R. 2015. Label-free and amplification-free miR-124 detection in human cells[J]. International Journal of Oncology,46(2):871-877.
Spidlen J,Breuer K,Rosenberg C,Kotecha N,Brinkman R R. 2012. FlowRepository:A resource of annotated flow cytometry datasets associated with peer-reviewed publications[J]. Cytometry (Part A),81(9):727-731.
Sun Y,Li Q,Gui H,Xu D P,Yang Y L,Su D F,Liu X. 2013. microRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines[J]. Cell Research,23(11):1270-1283.
Sun Y,Qin Z,Li Q,Wan J J,Cheng M H,Wang P Y,Su D F,Yu J G,Liu X. 2016. microRNA-124 negatively regulates LPS-induced TNF-alpha production in mouse macrophages by decreasing protein stability[J]. Acta Pharmacologica Sincia,37(7):889-897.
Vinayaka A C,Ngo T A,Kant K,Engelsmann P,Dave V P,Shahbazi M A,Wolff A,Bang D D. 2019. Rapid detection of Salmonella enterica in food samples by a novel approach with combination of sample concentration and direct PCR[J]. Biosensors & Bioelectronics,129:224-230.
(責任編輯 蘭宗寶)

