兩種生物除磷系統活性污泥中除磷菌的甄別——淀粉-缺氧/好氧交替與乙酸鹽-厭氧/好氧交替系統
周旭紅,袁林江*,陳 希,楊 睿,朱 淼,南亞萍,賀向峰,陳 勇
兩種生物除磷系統活性污泥中除磷菌的甄別——淀粉-缺氧/好氧交替與乙酸鹽-厭氧/好氧交替系統
周旭紅1,2,3,袁林江1,2,3*,陳 希4,楊 睿1,2,3,朱 淼1,2,3,南亞萍1,2,3,賀向峰1,2,3,陳 勇1,2,3
(1.西安建筑科技大學環境與市政工程學院,陜西 西安 710055;2.西安建筑科技大學西北水資源與環境生態教育部重點實驗室,陜西 西安 710055;3.西安建筑科技大學陜西省環境工程重點實驗室,陜西 西安 710055;4.西安工程大學城市規劃與市政工程學院,陜西 西安 710048)
為了直接識別出污泥中的聚磷細菌和其種屬,本研究采用4',6-二脒基-2-苯基吲哚(DAPI)染色和流式細胞熒光分選技術(FACS)對以淀粉為唯一碳源的缺氧/好氧序批式活性污泥(SBR)系統(R1)的缺氧末期和好氧末期以及以乙酸鹽為唯一碳源的厭氧/好氧SBR系統(R2)的好氧末期污泥的聚磷細菌進行了原位分選,并通過16S rRNA高通量測序技術鑒定了分選后細菌的種屬.結果表明,在R1中,缺氧期和好氧期均進行生物除磷,且缺氧期吸磷量大于好氧期. R2中發生著厭氧期釋磷、好氧期大量吸磷的傳統生物除磷.利用FACS在R1和R2污泥中均分選得到106個相對純度為85%的具有聚磷顆粒的細菌.測序結果表明,在R1系統中,缺氧段優勢的聚磷菌屬為(37.75%)、unclassified(14.15%)、(6.49%)、unclassified(0.027%)和(0.007%);好氧段優勢聚磷菌屬為(19.72%)、unclassified(14.62%)、(14.28%)、unclassified(0.046%)、unclassifiedGp3(0.036%)和(0.026%).R1系統中unclassified和僅僅在缺氧條件下具有聚磷功能,而unclassified、unclassifiedGp3和僅在好氧條件下才具有聚磷功能.在R2系統中,優勢聚磷菌群為(11.06%)、unclassified(9.29%)、unclassified(7.44%)、unclassified(7.34%)以及(0.31%).這意味著在新型的除磷系統(R1)中,參與除磷過程的細菌包括好氧,缺氧和兼性缺氧聚磷細菌,而在傳統的除磷系統(R2)中,參與除磷過程的細菌僅為好氧聚磷細菌.
聚磷菌;SBR;流式細胞熒光分選技術;高通量測序
早在20世紀70年代,人們已經明確認識到在厭氧 /好氧交替運行系統的活性污泥中會發生生物除磷:污水中的磷酸鹽濃度顯著降低、同時污泥的含磷率不斷上升的現象[1];也借助微生物染色觀察到污泥中有些細菌在厭氧條件下體內聚-β-羥丁酸(PHB)增加,聚磷顆粒(poly-P)減少,而在好氧條件下,這些細菌體內的PHB和聚磷顆粒含量變化情況恰好相反[2-3].但一直以來對這些聚磷的細菌“身份”沒有得到確認,只是概括性地將承擔生物除磷的細菌統稱為聚磷菌(PAOs),并按照其發生聚磷環境的不同將聚磷菌劃分為反硝化聚磷菌(DNPAOs)以及好氧聚磷菌兩大類[4-5].
盡管生物聚磷現象的發現至今已有半個多世紀,生物除磷也已被廣泛應用于城市污水處理中,但生物除磷活性污泥中聚磷菌到底是哪些種(屬)的細菌,仍尚無定論[6-7].1975年Fuhs等[8]最先從污水處理廠聚磷污泥中用平板劃線法分離出不動桿菌(),發現其能夠在生長培養基中大量吸收磷酸鹽,因而認定是污水處理廠污泥中主要聚磷菌,而Wagner等[9]認為不動桿菌并不是主要聚磷菌,因為分離得到的并沒有充分吸磷和釋磷:另有研究采用熒光抗體[10]或API系統[11]方法專門對實際污水處理廠污泥中的不動桿菌數量進行了追蹤,表明不動桿菌數量并不隨著系統除磷酸鹽能力提高而增加、兩者之間并沒有關聯性,不支持不動桿菌是主要聚磷菌的認識.1991年Nakamura等[12]從含磷率達到6%以上的污泥中劃線分離獲得了微球菌屬積磷小月菌(),該菌在厭氧條件下釋放磷酸鹽并吸收葡萄糖,與在活性污泥聚磷相似,但Santos等[13]研究發現并不儲存聚羥鏈烷酸(PHAs).1997年Stante等[14]在強化生物除磷系統(EBPR)中分離出俊片菌屬(),在培養基中具有厭氧釋磷和好氧吸磷行為,確定其為一種典型的聚磷細菌,但任世英等[15]研究發現只儲存少量磷酸鹽多聚物,且速度很慢.2007年Cai等[16]在實驗室活性污泥中純化分離出新型菌株GM6,該菌株在純化培養基具有較高的聚磷能力,但在厭氧/好氧交替環境中并沒有表現出典型的聚磷行為.2016年Terashima等[17]證實了日本氧化溝污水處理廠中脫氯單胞菌()為優勢菌屬,且分離得到純在培養時細胞內有多聚磷酸鹽.通過傳統平板劃線分離獲得純菌能夠在培養基中具有吸磷的菌還有:芽孢桿菌()[18-19]、葡萄球菌()[19],氣單胞菌()[20]等.但這些從聚磷污泥中分離得到的純菌大多沒有典型聚磷菌的代謝行為.有研究認為[21-22]聚磷菌要在有發酵產酸菌存在且與其它競爭水中揮發性脂肪酸(VFAs)的異養菌(如聚糖菌)共存下才表現出的適應性行為,因此分離純化后再鑒定可能無法通過其代謝行為來確定其聚磷菌身份.傳統平板劃線分離-培養法是對生物除磷活性污泥中細菌先進行分離鑒定,然后通過純菌的再培養條件下的代謝情況來判定某一個菌是否有聚磷菌的行為[23].但由于純培養所提供的條件與活性污泥混合菌群條件不同,因此分離得到的純菌的代謝過程和活性污泥中的細菌在實際環境中的代謝過程也可能會有所不同.這樣難免出現原本推測在活性污泥中可能具備聚磷能力的細菌卻在厭氧-好氧交替的純培養條件下不能呈現出釋磷-超量聚磷能力[17,24],同樣也會將某些隨著污泥聚磷能力增強,數量也隨之增加的細菌被推測為聚磷菌,然而這些細菌在經過分離培養后,很多是被否定的[25-26].
Kong等[27]通過熒光原位雜交(FISH)結合微自放射照相技術(MAR)顯示四球蟲菌()參與磷的攝取,被證實它是一種PAOs.Crocetti等[28]采用同樣方法確定了污泥中是污泥中的PAOs之一,開創了PAOs原位鑒定技術.但MAR-FISH技術原位鑒定技術步驟繁雜條件要求高.
FACS可將不同熒光標記細菌從混合細菌種直接“挑”出來,高效準確地實現目標細胞原位直接分離[29].2019年Wang等[30]應用流式細胞術細胞分選結合16S rRNA基因高通量測序甄別出生物除硫除磷系統中一些功能菌.該技術也可能成為PAOs原位分離的新技術.
目前鑒定出來除了在傳統厭氧/好氧交替環境中利用VFAs為碳源進行聚磷的微生物外,近年來還報道了在缺氧/好氧交替環境中利用淀粉為碳源聚磷的微生物.但對這種系統中聚磷菌的“身份”尚不清楚[31].因此,本文借助DAPI標記聚磷微生物并通過FACS對以淀粉為唯一碳源的缺氧/好氧生物除磷SBR中缺氧末端和好氧末端參與除磷的微生物,以及傳統以乙酸鈉為唯一碳源的SBR系統中聚磷菌進行了分選,對分選得到的所有聚磷菌經過高通量核酸測序后,確定了優勢聚磷菌的種類,為豐富聚磷菌的認識、探明生物除磷菌奠定了基礎.
1 材料與方法
1.1 反應器及運行條件
采用塑料材質的圓柱形反應器,柱體內徑18.7cm,柱高20cm,總有效容積4L.反應裝置如圖1所示,采用微孔曝氣器進行曝氣,曝氣量可由空氣流量計進行調節,氣體流速為200mL/min,自控裝置控制厭氧、缺氧、好氧等操作過程,厭氧和缺氧攪拌器轉速為300r/min.
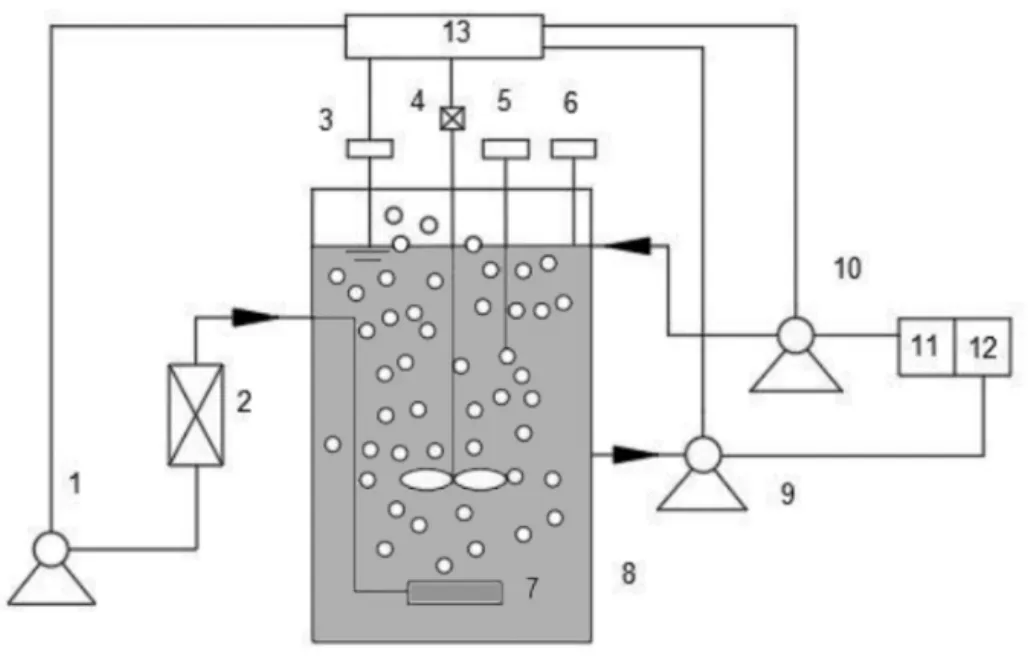
圖1 SBR系統實驗裝置
1.空氣泵;2.流量計;3.液位控制計;4.攪拌器;5.pH控制儀;6.DO測量儀;7.微孔曝氣頭;8.SBR反應器;9.出水泵;10.進水泵;11.進水箱;12.出水箱;13.可編程控制器
反應器接種污泥取自西安市某廢水處理廠厭氧/好氧(A/O)系統,取回的污泥過篩(50目)后直接加入R1和R2系統中馴化培養.R1系統每天運行4個周期,每周期6h.其中,進水8min,缺氧90min,好氧210min,沉淀40min,排水加閑置12min.R2系統運行周期和時間安排與R1系統一致,其中,厭氧90min. R1、R2兩系統水力停留時間均為12h,污泥停留時間均控制在15d左右,均保持污泥濃度在3000~ 3500mg/L之間,好氧期溶解氧(DO)也均保持在2mg/L左右.
1.2 進水水質
采用人工配水模擬生活廢水,R1反應器配方如下(g/L): MgSO4·7H2O:0.1,無水 CaCl2:0.01, NaNO3: 0.122,淀粉:0.4,NH4Cl:0.038;R2反應器配方如下(g/L): 無水CaCl2:0.01,NaAc:0.51, MgSO4·7H2O:0.1, NH4Cl:0.0608,其中每升配水加入微量元素濃縮液0.5mL,微量元素濃縮液組成如下(g/L): Na2MoO4·
H2O:0.06,FeSO4·7H2O:1.54,CuSO4·5H2O:0.03,CoCl2·7H2O:0.15,KI:0.18,H3BO3:0.15,MnCl2·4H2O:0.12,
ZnSO4·7H2O:0.12,EDTA:10.進水的pH值采用鹽酸和氫氧化鈉調節,控制范圍為7.3~7.5.
1.3 水質參數分析
試驗過程中按照每個周期的設定時間定期取樣,從反應器最上部取混合液,取出的樣品用濾紙過濾后用于測定水質指標.COD、TP和PO43--P采用標準方法測定[32],采用PE680型氣相色譜儀測定PHB和聚β-羥戊酸(PHV)[33],測定VFAs和乳酸采用安捷倫氣相色譜儀(hp 6890N,Amercian)型,DO和pH值的測定分別采用HACH-HQ40d型溶解氧儀和PB-20型pH計,微生物特征分析采用奧林巴斯(Olympus)型熒光顯微鏡進行觀察.
1.4 活性污泥DAPI染色
分別取R1、R2反應器不同階段的活性污泥用DAPI進行染色.染色具體步驟同文獻[34-35]采用的方法.通過染色發現R1缺氧階段與好氧階段都有聚磷顆粒形成,而R2系統厭氧末端并未發現聚磷顆粒的存在,但好氧末端有大量聚磷顆粒的合成.因此認為R1中缺氧階段和好氧階段具有微生物聚磷,參與R2除磷的微生物僅在好氧期吸磷.選取染色后有聚磷顆粒的樣本將其制備成菌懸液并分選其中能夠聚磷的微生物.
1.5 流式細胞術收集含poly-P的細胞
DAPI染色方法和FACS分選方法在文獻[36-37]的方法上進行改進.具體為:
(I)菌懸液制備與染色:取運行穩定的R1反應器缺氧末端和好氧末端活性污泥和R2反應器好氧末端的活性污泥各5mL于50mL離心管中,先離心5min(4℃, 10000r/min),傾去上清液后在離心管中加入10mL 0.1′磷酸鹽緩沖溶液(PBS),重復離心,棄上清3次,去除活性污泥表面雜質.最后加0.1×PBS緩沖液至30mL,用超聲波細胞粉碎機(型號:BILON92-II)將細胞混合液離散(單位體積輸入能量:72.5J/mL).所制備的菌懸液中值粒徑為(30±1.27)μm,吸光度(OD)值在0.4~0.5之間,脫氧核糖核酸(DNA)含量僅為3.44mg/(gVSS).制好的菌懸液每毫升加入30μL的甲酸,將pH值調至6.5以下,以抑制微生物代謝.用于菌懸液染色的DAPI濃度為10μg/mL,每毫升菌懸液加入10μL的DAPI染色液,37℃避光震蕩染色30min.流式細胞分選前需用細菌渦旋儀(UVS-1)將菌懸液渦旋30s.
(Ⅱ)流式細胞儀分選細菌:采用北京易科生物科技攀博有限公司的流式細胞分選儀(BD AriaⅢ Special Order)進行細菌分離.DAPI用355nm(UV)激光器激發,并用460/50nm帶通濾光片收集標準DAPI-DNA發射光;用585/15nm帶通濾波片收集了黃色的DAPI-poly-P發射光.DAPI-DNA和DAPI- poly-P的測量值以對數標度進行采集,并使用Flow Jo軟件7.6.5版進行采集后分析.分選速率為2000個/s,設置四路分選中的左二通道電壓使液滴偏轉到載玻片上,用于熒光顯微鏡觀察分選純度,其余偏轉至無菌離心管中.通過熒光顯微鏡觀察后分選目標菌群的純度高達85%以上.
1.6 流式細胞術后分選得到的菌16S rDNA基因測序鑒定
取R1反應器污泥混合液以及兩系統經過流式細胞儀分選后的所有細菌菌群進行16S rDNA高通量測序(上海生物工程公司).所測序菌群的擴增引物選擇Illumina Miseq測序平臺所提供的V3-V4區域的通用引物341F(5'-CCTACGGGNGGCWGCAG-3')和805R(5'-GACTACHVGGGTATCTAATCC-3').
2 結果與分析
2.1 SBR系統的除磷特性
如表1、2所示,在R1中,整個周期內無釋磷現象發生,淀粉的主要水解產物為乳酸,系統缺氧期中基本沒有PHB和PHV的合成,微生物合成內碳源主要為糖原.而R2系統相比R1有厭氧末端合成較多PHB和PHV,表現為典型傳統厭氧釋磷、好氧吸磷的除磷模式.總之,R1存在著不同于傳統除磷的過程,推測參與R1系統除磷微生物可能也不同于傳統除磷體系,此結論與Luo等[31]研究相符.
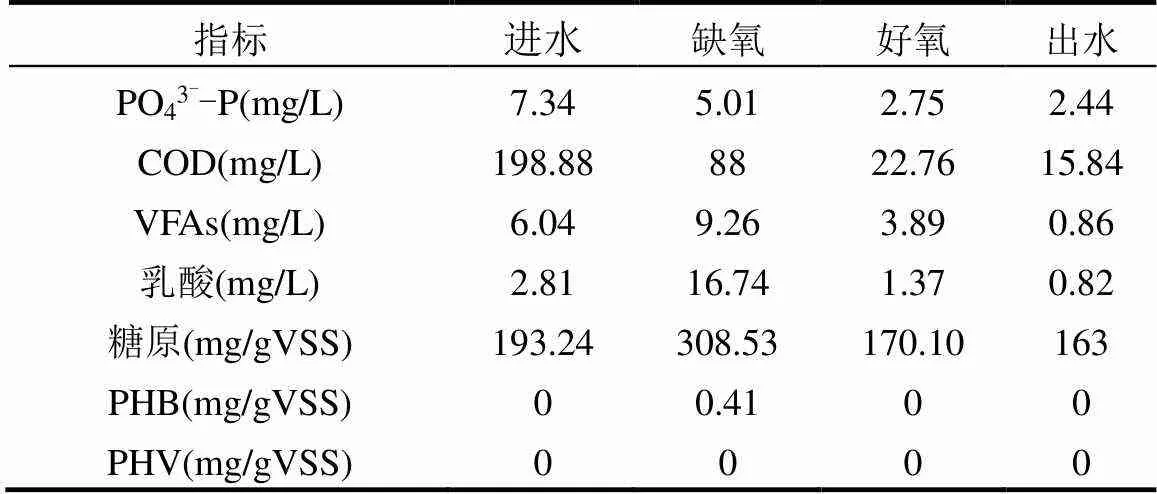
表1 R1反應器運行中系統周期內各物質濃度變化
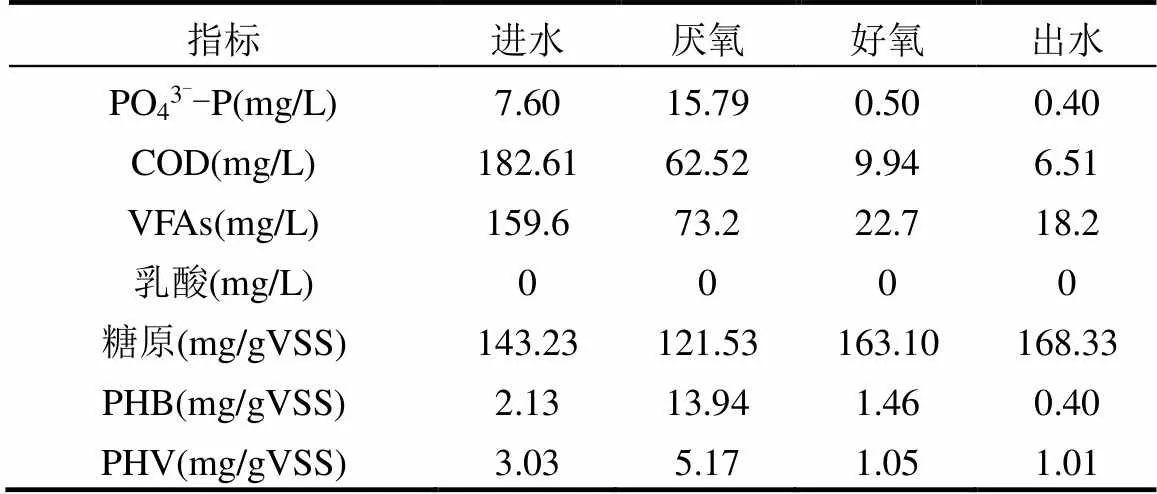
表2 R2反應器運行中系統周期內各物質濃度變化
2.2 流式細胞儀分選系統中的PAOs
借助FACS分選R1系統同周期內缺氧末端、好氧末端以及R2好氧末端的除磷微生物.如圖2所示,首先排除細菌中的雜質和細菌碎片,接著除去細菌中的粘結細菌后剩余游離細菌,游離細菌被DAPI染料染色后,選取能夠發出DAPI-DNA熒光的細菌,最后在所有具備DAPI-DNA熒光的細菌中選取出能夠發出DAPI-poly-P熒光的細菌,即為目標細菌.
為了獲得用于產生16S rRNA基因克隆文庫的足夠的DNA,所取每個樣品各準確分選106個目標細菌.R1缺氧末端和好氧末端用于分選的細菌總數分別為157435053和8426607,而R2好氧末端所用細菌總數為6429640,從分選所用細菌數量占比可以看出具備聚磷功能的微生物在R1、R2兩個除磷系統中均為少部分.
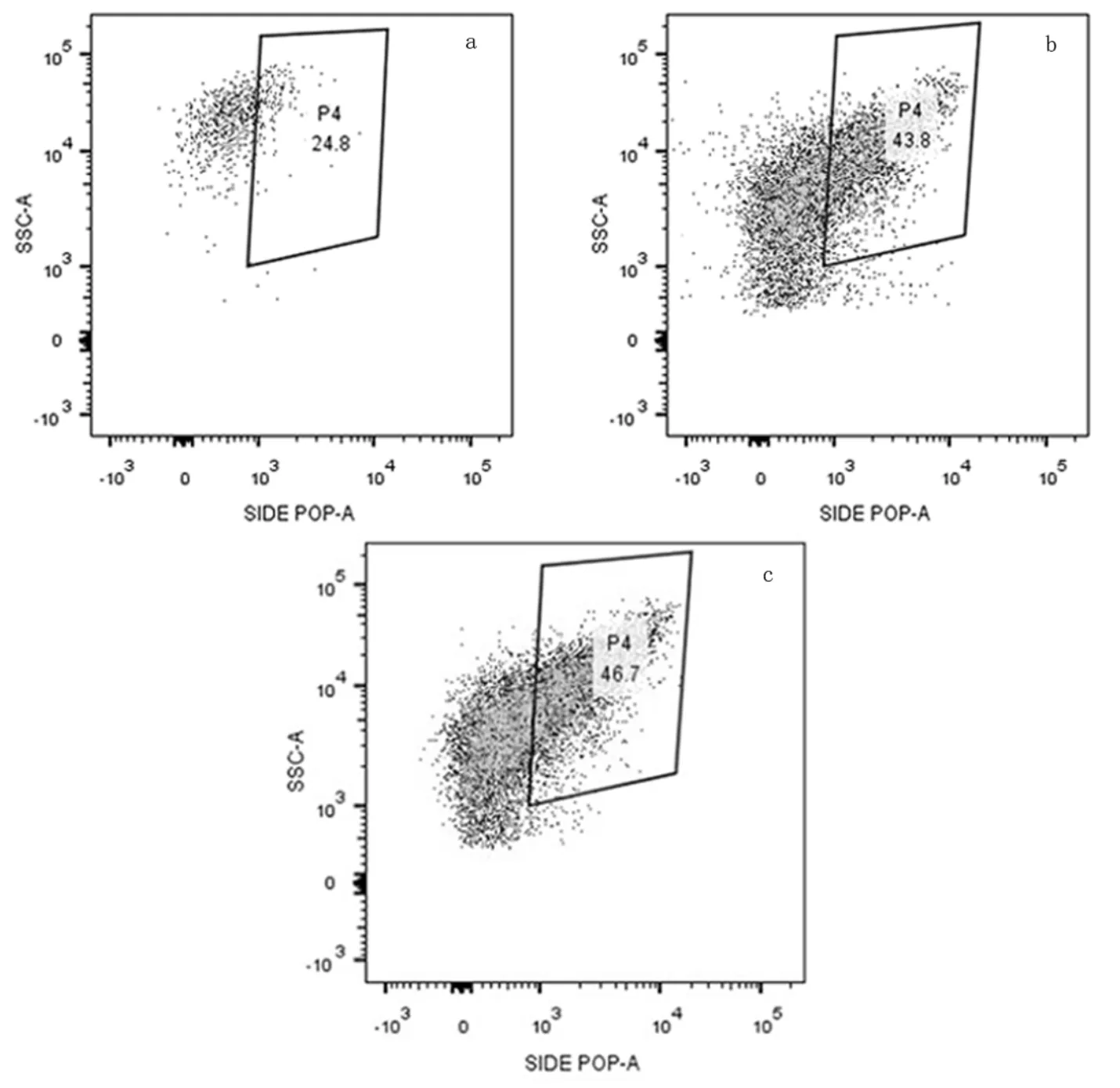
圖2 流式細胞儀分選R1中缺氧末端、好氧末端和R2中好氧末端具備聚磷顆粒細菌的檢測結果
a. R1缺氧末;b. R1好氧末端;c. R2好氧末端;不同點在相應區域分布的密集情況代表不同細菌于相應區域分布的密集情況
2.3 微生物菌群分析
如圖3所示,在R1系統中,分選前優勢菌屬為(21.56%)和(15.27%).經過FACS分選后,缺氧末端和好氧末端活性污泥中均沒有檢測到,說明此細菌在R1中并沒有參與聚磷,因此體內沒有聚磷顆粒形成.在分選后的缺氧期聚磷菌和好氧期聚磷菌中占比分別為6.49%和14.28%,顯然,在R1系統中,在缺氧期和好氧期均發揮了一定的聚磷作用.2008年任世英等[38]從海洋中分離得到,并證明其除磷能力,且能夠水解淀粉.本研究的缺氧/好氧系統中,分選前僅占0.019%,在缺氧末端和好氧末端聚磷菌中此菌占比分別高達37.75%和19.72%.說明在以淀粉為唯一碳源的缺氧/好氧系統中,主要起聚磷作用的優勢菌群為.此外unclassified分選前豐度僅為0.08%,經過分選后,在R1缺氧末端和好氧末端除磷菌中,unclassified豐度分別為14.15%和14.62%,可見unclassified在缺氧期和好氧期均發揮了重要的聚磷作用.(0.007%)以及unclassified(0.027%)等細菌僅在缺氧期具備聚磷能力,而unclassified0.046%)、unclassified(0.036%)和(0.026%)等細菌僅在好氧期具有聚磷能力.這說明細菌除磷與環境息息相關,聚磷只有在適宜的條件下才能進行,缺氧期與好氧期除磷機制不同[31,34],發揮除磷作用的細菌也不同,因此具有聚磷顆粒的細菌也不同.
在R2系統中,(11.06%)、unclassified(9.29%)和unclassified(7.44%) unclassified(7.34%)等為主要參與除磷的微生物.其中是最近兩年新確認的同一樣重要的聚磷菌,與也有一些共同的特征,并且生態生理學與非常相似[17].
鑒定結果表明,unclassified為R1、R2兩個系統共有的聚磷菌.在R1系統中,unclassified在分選前后占比均不到0.35%,這說明在此系統中并不起主導除磷的作用.而此菌在分選后的R2系統中占比為3.69%,可見unclassified在R2系統中的占比高于R1,這是由于unclassified更傾向于在以短鏈脂肪酸乙酸鈉為碳源的系統中聚磷[39].

圖3 R1系統分選前后及R2系統分選后微生物屬水平優勢菌屬的相對豐度
X2、X3的優勢菌群中已排除X1中不存在的細菌;X1:R1分選前;X2:R1缺氧末分選后;X3:R1好氧末分選后;X4:R2好氧末分選后
相比兩個系統中主要的除磷優勢菌群鑒定結果,雖然兩種系統的反應時間相同,但由于運行條件和基質不同,主要參與除磷的微生物的種類和數量也存在較大差異.此外既能夠在缺氧的環境中聚磷,也能在好氧環境中聚磷,目前尚未有人報道該菌屬在反應器活性污泥中的聚磷能力,研究通過DAPI標記聚磷顆粒細菌后利用FACS技術分選發現其在活性污泥中的聚磷能力為豐富聚磷菌的認識、探明生物除磷菌奠定了基礎.
3 結論
3.1 在以淀粉為唯一碳源的缺氧/好氧系統中,實現了缺氧期和好氧期均吸磷,并且缺氧期的吸磷量大于好氧期吸磷量,而以乙酸鈉為唯一碳源的系統中為傳統除磷過程.
3.2 在R1系統中,缺氧期和好氧期優勢聚磷菌為、和unclassified等,其中、和僅在缺氧期具備聚磷功能,unclassifiedGp3、和也僅在好氧期才會發揮聚磷能力.在R2系統中優勢聚磷菌為unclassified、unclassified、unclassified以及.
3.3 在新型的除磷系統(R1)中,參與除磷過程的細菌包括缺氧、好氧和兼性缺氧聚磷細菌,表明了該系統除磷由多種不同類型的除磷菌承擔,并在多種環境下都出現磷的攝取;而在傳統除磷系統(R2)中,參與除磷過程的細菌僅有好氧聚磷細菌.
[1] Nicholls H A, Osborn D W. Bacterial stress: prerequisite for biological removal of phosphorus [J]. Water Pollution Control Federation, 1979,51(3):557-569.
[2] Dorofeev A G, Nikolaev Y A, Mardanov A V, et al. Role of phosphateaccumulating bacteria in biological phosphorus removal from wastewater [J]. Applied Biochemistry and Microbiology, 2020, 56(1):1-14.
[3] 王亞東,王少坡,鄭莎莎,等.生物除磷系統的聚磷微生物種群及其檢測方法[J]. 環境工程, 2015,24(2):21-26.
Wang Y D, Wang S P, Zheng S S, et al Phosphorus accumulating microorganism population of biological phosphorus removal system and its detection method [J] Environmental engineering, 2015,24(2): 21-26.
[4] Rao N N, Gómez-García M R, Kornberg A. Inorganic polyphosphate: essential for growth and survival [J]. Annual review of biochemistry, 2009,78:605-647.
[5] 李 冬,李曉瑩,楊 杰,等.后置缺氧SBR短程反硝化除磷[J]. 中國環境科學, 2017,37(8):2994-3001.
Li D, Li X Y, Yang J, et al. Post-hypoxic SBR short-range denitrification for phosphorus removal [J]. China Environmental Science, 2017,37(8):2994-3001.
[6] Nielsen P H, McIlroy S J, Albertsen M, et al. Re-evaluating the microbiology of the enhanced biological phosphorus removal process [J]. Current opinion in biotechnology, 2019,57:111-118.
[7] Qiu G, Zuniga-Montanez R, Law Y, et al. Polyphosphate- accumulating organisms in full-scale tropical wastewater treatment plants use diverse carbon sources [J]. Water research, 2019,149:496- 510.
[8] Fuhs G W, Chen M. Microbiological basis of phosphate removal in the activated sludge process for the treatment of wastewater [J]. Microbial Ecology, 1975,2(2):119-138.
[9] Wagner M, Erhart R, Manz W, et al. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge [J]. Applied and Environmental Microbiology, 1994,60(3):792- 800.
[10] Cloete T E, Steyn P L. The role of Acinetobacter as a phosphorus removing agent in activated sludge [J]. Water Research, 1988,22(8): 971-976.
[11] Meganck M, Malnou D, Le Flohic P, et al. The importance of the acidogenic microflora in biological phosphorus removal [J]. Water science and technology, 1985,17(11/12):199-212.
[12] Nakamura K, Masuda K, Mikami E. Isolation of a new type of polyphosphate accumulating bacterium and its phosphate removal characteristics [J]. Journal of fermentation and bioengineering, 1991, 71(4):258-263.
[13] Santos M M, Lemos P C, Reis M A M, et al. Glucose metabolism and kinetics of phosphorus removal by the fermentative bacterium Microlunatus phosphovorus [J]. Applied and Environmental Microbiology, 1999,65(9):3920-3928.
[14] Stante L, Cellamare C M, Malaspina F, et al. Biological phosphorus removal by pure culture of Lampropedia spp [J]. Water research, 1997,31(6):1317-1324.
[15] 任世英,肖 天.聚磷菌體內多聚物的染色方法[J]. 海洋科學, 2005, 29(1):59-63.
Ren S Y, Xiao T. Staining method of polymer in polyphosphate accumulating bacteria [J]. Marine science, 2005,29(1):59-63.
[16] Cai T M , Guan L B , Chen L W , et al. Enhanced biological phosphorus removal with P seudomonas putida GM6 from activated sludge [J]. Pedosphere, 2007,17(5):624-629.
[17] Terashima M, Yama A, Sato M, et al. Culture-Dependent and -Independent Identification of Polyphosphate-Accumulating Dechloromonas spp. Predominating in a Full-Scale Oxidation Ditch Wastewater Treatment Plant [J]. Microbes and Environments, 2016, 31(4):449-455.
[18] 周明璟,紀樹蘭,崔丹紅,等.厭氧/好氧交替快速篩選聚磷菌及其生理特性的研究[J]. 中國環境科學, 2012,32(10):1838-1844.
Zhou M J, Ji S L, Cui D H, et al. Rapid Screening of Phosphorus Accumulating Bacteria and Their Physiological Characteristics by Alternating Anaerobic/Aerobic [J]. China Environmental Science, 2012,32(10):1838-1844.
[19] 張立成,李艷美,袁雅姝,等.亞硝化反硝化聚磷菌的篩選及生化特性分析[J]. 中國給水排水, 2013,29(11):77-80.
Zhang L C, Li Y M, Yuan Y S, et al. Screening and biochemical characteristic analysis of nitrosating and denitrifying phosphorus accumulating bacteria [J]. China Water Supply and Drainage, 2013, 29(11):77-80.
[20] 李 慧,劉丹丹,陳文清.反硝化聚磷菌的篩選及脫氮除磷特性[J]. 環境工程, 2016,34(4):25-28.
Li H, Liu D D, Chen W Q. Screening of denitrifying phosphorus- accumulating bacteria and their characteristics of nitrogen and phosphorus removal [J]. Environmental Engineering, 2016,34(4):25- 28.
[21] 郝曉地,陳 嶠,劉然彬.Tetrasphaera聚磷菌研究進展及其除磷能力辨析[J]. 環境科學學報, 2020,40(3):741-753.
Hao X D, Chen Q, Liu R B. Research Progress of Tetrasphaera Phosphorus Accumulating Bacteria and Analysis of Phosphorus Removal Ability [J]. Journal of Environmental Science, 2020,40(3): 741-753.
[22] Qian W, Linjiang Y, Xi C, et al. Biological phosphorus removal and its mechanism in anoxic/aerobic continuous flow system with different carbon sources [J]. Chinese Journal of Environmental Engineering, 2021,15(3):954-961.
[23] 馬 放,楊菲菲,李 昂,等.1株高效反硝化聚磷菌的生物學特性研究[J]. 環境科學, 2011,32(9):2710-2715.
Ma F, Yang F F, Li A, et al. Biological characteristics of a high- efficiency denitrifying phosphorus accumulating bacteria [J]. Environmental Science, 2011,32(9):2710-2715.
[24] Sun L, Zhao X, Zhang H, et al. Biological characteristics of a denitrifying phosphorus-accumulating bacterium [J]. Ecological engineering, 2015,81(2015):82-88.
[25] Kong Y, Nielsen J L, Nielsen P H. Identity and ecophysiology of uncultured actinobacterial polyphosphate-accumulating organisms in full-scale enhanced biological phosphorus removal plants [J]. Applied and environmental microbiology, 2005,71(7):4076-4085.
[26] 蔣志云,韋佳敏,繆新年,等.ABR-MBR工藝反硝化除磷微生物群落特征分析[J]. 環境工程學報, 2019,13(7):1653-1661.
Jiang Z Y, Wei J M, Miao X N, et al. Characteristic analysis of microbial community for denitrification and phosphorus removal by ABR-MBR process [J]. Chinese Journal of Environmental Engineering, 2019,13(7):1653-1661.
[27] Kong Y, Xia Y, Nielsen P H. Activity and identity of fermenting microorganisms in full-scale biological nutrient removing wastewater treatment plants [J]. Environmental microbiology, 2008,10(8):2008- 2019.
[28] Crocetti G R, Hugenholtz P, Bond P L, et al. Identification of polyphosphate-accumulating organisms and design of 16S rRNAdirected probes for their detection and quantitation [J]. Applied and environmental microbiology, 2000,66(3):1175-1182.
[29] Hammes F, Berney M, Wang Y, et al. Flow-cytometric total bacterial cell counts as a descriptive microbiological parameter for drinking water treatment processes [J]. Water research, 2008,42(1/2):269-277.
[30] Wang H G, Huang H, Liu R L, et al. Investigation on polyphosphate accumulation in the sulfur transformation-centric EBPR (SEBPR) process for treatment of high-temperature saline wastewater [J]. Water research, 2019,167:115138.
[31] Luo D, Yuan L, Liu L, et al. Biological phosphorus removal in anoxic-aerobic sequencing batch reactor with starch as sole carbon source [J]. Water Science and Technology, 2017,75(1):28-38.
[32] 國家環保局本書編委會.水和廢水監測分析方法[M]. 北京:水和廢水監測分析方法, 1989.
The State Environmental Protection Administration's book editorial committee. Water and wastewater monitoring and analysis methods [M]. Beijing. Water and wastewater monitoring and analysis methods, 1989.
[33] 李夕耀,彭永臻,王淑瑩,等.聚磷菌胞內多聚物的分析檢測方法 [J]. 四川環境, 2009,28(2):106-111.
Li X Y, Peng Y Z, Wang S Y, et al. Analysis and detection method of intracellular polymers of phosphorus accumulating bacteria [J]. Sichuan Environment, 2009,28(2):106-111.
[34] Huo X, Yuan L, Wang Q, et al. Nitrate Promotes Phosphorus Removal in the Anoxic–Aerobic Sequencing Batch Reactor with Starch as Sole Carbon Source [J]. Environmental Engineering Science, 2021,38(2): 66-73.
[35] 葛艷輝,趙 林,周 艷.聚磷菌胞內聚合物的染色條件優化及染色方法比較[J]. 環境科學與技術, 2014,37(2):1-6.
Ge Y H, Zhao L, Zhou Y. Optimization of dyeing conditions and comparison of dyeing methods for intracellular polymers of phosphorus accumulating bacteria [J]. Environmental Science and Technology, 2014,37(2):1-6.
[36] Wang H G, Biswal B K, Mao Y P, et al. Multiple-cycle operation of sulphur-cycle-enhanced biological phosphorus removal to maintain stable performance at high temperatures [J]. Bioresource technology, 2019,289:121736.
[37] Terashima M, Kamagata Y, Kato S. Rapid enrichment and isolation of polyphosphate accumulating organisms through 4’6-diamidino-2- phenylindole (DAPI) staining with fluorescence-activated cell sorting (FACS) [J]. Frontiers in microbiology, 2020,11(2015):793.
[38] 任世英,張宇紅,張武昌,等.海洋聚磷菌Halomonas YSR-3的除磷特性研究[J]. 高技術通訊, 2008,18(7):743-747.
Ren S Y, Zhang Y H, Zhang W C, et al. Study on Phosphorus Removal Characteristics of Marine Phosphorus Accumulating Bacteria Halomonas YSR-3 [J]. High-Tech Communications, 2008,18(7):743- 747.
[39] Miyauchi R, Oki K, Aoi Y, et al. Diversity of nitrite reductase genes in “Candidatus Accumulibacter phosphatis”-dominated cultures enriched by flow-cytometric sorting [J]. Applied and environmental microbiology, 2007,73(16):5331-5337.
Screening of phosphorus removing bacteria from activated sludge for biological phosphorus removal: starch-anoxic/aerobic alternation and acetate-anaerobic/aerobic alternation system.
ZHOU Xu-hong1,2,3, YUAN Lin-jiang1,2,3*, CHEN Xi4, YANG Rui1,2,3, ZHU Miao1,2,3, NAN Ya-ping1,2,3, HE Xiang-feng1,2,3, CHEN Yong1,2,3
(1.School of Environmental and Municipal Engineering, Xi’an University of Architecture and Technology, Xi’an 710055, China;2.Key Laboratory of Northwest Water Resource, Environment and Ecology, Ministry of Education, Xi’an University of Architecture and Technology, Xi’an 710055, China;3.Shaanxi Key Laboratory of Environmental Engineering, Xi’an University of Architecture and Technology, Xi’an 710055, China;4.School of Urban Planning and Municipal Engineering, Xi'an University of Engineering, Xi'an 710048, China)., 2022.42(9):4166~4173
In order to identify phosphorus accumulating organisms(PAOs) in situ from activated sludge. 4', 6-diamidino-2- phenylindole (DAPI) staining and flow cytometry fluorescence sorting (FACS) were used to sceen the PAOs from the two sludge at the anoxic/aerobic SBR system (R1) with starch as the only carbon source and the anaerobic/aerobic system (R2) with acetate as the only carbon source. The species of the sorted bacteria were identified by 16S rRNA high-throughput sequencing. The results showed that in the R1, biological phosphorus removal was carried out in both the anoxic periods and aerobic periods. The phosphorus uptake in the anoxic period was greater than that in the aerobic period. In the R2, the phosphorus release in the anaerobic period and a large amount of phosphorus absorption in the aerobic period occurs. Results of the in situ fluorescence staining showed that 106bacteria with a relative purity of 85% with phosphorus-accumlating cell were sorted from the R1 and the R2. The sequencing results showed that in the R1, the dominant genera of PAOs in the anoxic periods were(37.75%), unclassified(14.15%),(6.49%), unclassified(0.027%) and(0.007%). The dominant PAOs in the aerobic periods were(19.72%), unclassified(14.62%),(14.28%), unclassified(0.046%), unclassifiedGp3 (0.036%) and(0.026%). In the R1, unclassifiedandonly had phosphorus accumulation function under anoxic periods, while unclassified, unclassifiedGp3 andonly had phosphorus accumulation function under aerobic periods. In the R2, the dominant PAOs were(11.06%), unclassified(9.29%), unclassified(7.44%), unclassified(7.34%) and(0.31%). This means that in the R1, the bacteria involved in the phosphorus removal process include aerobic, anoxic and both aerobic and anoxic phosphorate accumulating bacteria three types. In the R2, the bacteria involved in the phosphorus removal process are only aerobic phosphorus accumulating bacteria.
phosphorus accumulating bacteria;SBR;flow cytometry fluorescence sorting technology;high-throughput sequencing
X703.5
A
1000-6923(2022)09-4166-08
2022-02-10
國家自然科學基金資助項目(51078304,51278406);陜西省自然科學基金資助項目(2021JQ-690)
*責任作者, 教授, yuanlinjiang@xauat.edu.cn
周旭紅(1996-),女,甘肅定西人,西安建筑科技大學碩士研究生,主要從事廢水生物處理理論與技術研究.

