Atoh1基因與POU4F3、Gfi1協同作用促毛細胞再生的研究進展*
何才蓉, 楊影, 黃金煒, 陳鳳嬌, 陸瑩, 丁潔△
基因與POU4F3、Gfi1協同作用促毛細胞再生的研究進展*
何才蓉1, 楊影1, 黃金煒1, 陳鳳嬌2, 陸瑩3, 丁潔1△
(1貴州大學生命科學學院/農業生物工程研究院,山地植物資源保護與種質創新教育部重點實驗室,貴州 貴陽 550025;2貴州省人民醫院,貴州 貴陽 550002;3貴州中醫藥大學基礎醫學院解剖學教研室,貴州 貴陽 550025)
毛細胞再生;直接重編程;互作機制;聽力損失
聽力損失是導致語言障礙和影響人口素質的一種世界性頑疾,主要由內耳毛細胞不可逆損傷所致。治療此類頑疾的核心是毛細胞再生。直接重編程作為一種細胞再生技術,無需經過多能性細胞階段便可將一種終末分化細胞直接轉變為其他類型的功能細胞或祖細胞,因此也稱為譜系重編程[1-2],包括直接和間接兩種方式。直接的方式是通過導入譜系特異性轉錄因子誘導一種類型終末分化細胞轉化為另一種細胞類型,如慢病毒載體攜帶轉錄因子編碼基因感染支持細胞或成纖維細胞,經細胞形態、相關基因和蛋白表達等參數驗證獲得與正常內耳毛細胞相似的細胞,從而實現毛細胞再生的目的[3-5]。
是內耳毛細胞發育和再生的關鍵調節因子。研究表明,雖然基因能夠將耳蝸非感覺細胞直接重編程為新生毛細胞,但其重編程能力具有年齡依賴性[6]。為了規避這種情況,探索簡單有效的譜系特異性轉錄因子組合可顯著促進毛細胞分化、成熟和重編程效率,是毛細胞再生和功能重塑的可行方案。在眾多的轉錄因子中,POU結構域第4類轉錄因子3(POU domain class 4 transcription factor 3,Pou4f3)和獨立生長因子1(growth factor independent-1,Gfi1)可與共表達促進生成功能成熟的毛細胞而備受關注。已有研究表明,利用慢病毒在人類成纖維細胞培養物中過表達、和(簡稱)可促人類體細胞直接重編程為表達毛細胞標志物的細胞[7]。因此利用直接重編程技術獲取人類內耳毛細胞將成為一個新的研究方向。但三者在毛細胞再生及功能重塑中的作用、上下游毛細胞發育相關基因及互作機制未有系統闡明。本文就Atoh1、Pou4f3和Gfi1在毛細胞再生和功能重塑中的調控作用及互作機制的最新研究報道進行綜合整理,匯總與毛細胞發育相關的信號通路和關鍵基因,闡述三者間的表達調控方式,為未來使用Atoh1-Pou4f3-Gfi1直接重編程策略治療聽力損失提供參考資料。
1 Atoh1在脊椎動物內耳中的表達和功能
1.1Atoh1的結構和功能,又名、或,位于染色體4q22,全長1.06 kb,含一個外顯子。轉錄因子Atoh1屬第Ⅱ類堿性螺旋-環-螺旋(basic helix-loop-helix, bHLH)轉錄因子,可與第Ⅰ類bHLH轉錄因子如E47結合形成異二聚體,直接與CAGCTG和CAGGTG等E-box基序結合以激活下游靶基因的轉錄。含有較高比例的脯氨酸殘基,可參與蛋白質互作,C端的高比例絲氨酸殘基意味著的功能可能受這些絲氨酸殘基的磷酸化調節。此外,參與多種細胞類型的發育,敲除的小鼠缺乏腸道上皮細胞,小腦和脊髓神經發育不完全,內耳毛細胞完全缺失[8]。的表達和激活與腫瘤的發生、發展和轉移有關[9]。已證實在結腸和皮膚中充當腫瘤抑制基因[10],在髓母細胞瘤中作為癌基因起作用[11],表明其在不同細胞類型的分化和增殖中的多重作用。由于在耳蝸感覺上皮及非感覺上皮過表達可誘導原位及異位毛細胞再生的能力使得成為當前研究的熱點。
1.2在耳蝸中的發育表達模式的表達時序是毛細胞分化和存活的關鍵,決定毛細胞分化的特異性。在小鼠胚胎發育期間,最初是在耳蝸底部附近內耳祖細胞中檢測到的,隨后在 E13.5~E17.5穩定增加并沿頂部擴散[12]。在胚胎后期,多表達于毛細胞中以啟動毛細胞分化程序,在支持細胞內僅有少量表達且處于抑制狀態,而在非感覺細胞內則完全沒有[13]。毛細胞發育過程中的功能存在兩個關鍵時期,在E15.5~E17.5 期間缺失會導致毛細胞快速死亡,而在E17.5后缺失會破壞維持聽覺功能所需的毛束結構以致毛細胞延遲死亡。由此可見,是毛細胞正常分化、存活和成熟的必要條件。出生后一周,表達量開始下調,遵循從耳蝸管的基部向頂部的梯度,隨著聽覺毛細胞成熟而關閉[14]。
1.3在毛細胞再生中的作用是毛細胞形態和功能再生過程中必不可少的基因。基因敲除鼠的耳蝸和前庭感覺毛細胞完全缺失,大多數支持細胞標志基因的表達顯著降低[15]。提示的丟失不僅直接影響毛細胞存活,還會破壞周圍支持細胞的發育,間接控制感覺上皮鑲嵌發育。內耳毛細胞和支持細胞來源于相同前體細胞,故通常采用上調在耳蝸支持細胞中的表達水平以促進支持細胞向毛細胞的轉化[3],但新生毛細胞仍不成熟,無法長期存活。在支持細胞中的重編程能力隨年齡增長而受到限制[6],說明僅過表達不足以有效且持續地實現毛細胞功能成熟。毛細胞的形成和成熟受多種信號通路和轉錄因子的時空調控,作為毛細胞分化及存活過程中的先鋒因子,其功能的發揮依賴與其他因子的協同作用。
1.4結合其他轉錄因子的毛細胞重編程策略已知有多種信號通路參與調節表達,如Notch信號通路、Shh信號通路、Wnt信號通路、FGF信號通路和BMP信號通路等。此外β-連環蛋白(β-catenin、E2F轉錄因子1(E2F transcription factor 1,)、GATA結合蛋白3(GATA binding protein-3,Gata3)等轉錄因子可增強的表達[17-18],癌高甲基化基因1(hypermethylated in cancer 1,Hic1)、Zic家族成員1(Zic family member 1,Zic1)則抑制了Atoh1活性[19-20]。性別決定相關基因簇2(sex determining region Y,Sox2)、同源異型盒基因1(sineoculishomeobox homolog1,Six1)、Gata3等轉錄因子位于Atoh1上游。神經元素1 (neurogenin1, Neurog1)和Atoh1通過神經源分化因子1(neurogenic differentiation factor1, Neurod1)介導互為拮抗關系,在神經元與毛細胞的命運決定中發揮作用[4, 21]。Sox2表達于整個感覺上皮細胞,通過激活Neurog1、Neurod1、分化抑制因子(inhibitor of differentiation,Id)1-3和Hes/Hey家族等因子以抑制的表達。上皮祖細胞分化為毛細胞后,Atoh1的表達導致Sox2下調,因此Atoh1 mRNA激活后可下調Sox2以維持Atoh1在毛細胞發育中的表達水平,Sox2的下調由Six1介導[22]。如圖1所示,在毛細胞發育過程中Shh信號作為Atoh1的負性調控因子,通過FGF信號通路介導維持Hey1和Hey2表達水平,從而負調節Atoh1以防止毛細胞過早分化[23]。因此,調控Shh信號-FGF信號-Hey1/Hey2-Atoh1這一分子通路可能是誘導毛細胞再生的潛在途徑。此外,Atoh1受分化抑制基因Id1-3的負調控[24]。在耳蝸中Id蛋白受BMP信號調節,并可能受到細胞周期蛋白依賴性激酶2(cyclin-dependent kinase 2, Cdk-2)的負調控[25]。Atoh1表達后,其下游靶基因如Gfi1、Pou4f3和Barhl1對毛細胞的成熟和長期存活也是必要的。由此可見,在內耳發育的不同階段調控上述關鍵因子和信號通路對毛細胞的正常發育至關重要,有助于為毛細胞再生直接重編程技術制定可行方案。如圖2所示,幾種關鍵重編程轉錄因子與Atoh1的聯合調控可顯著促進毛細胞再生效率:Walters等[16]驗證,Atoh1過表達結合細胞周期蛋白激酶抑制因子(cyclin dependent kinase inhibitor 1, p27kip1)缺失或Atoh1和Gata3、Pou4f3的聯合激活足以將成年小鼠的支持細胞轉化為毛細胞,并克服轉分化的年齡依賴性。Ahmed等[26]鑒定眼發育缺失1(eyes absent1, Eya1)、Six1和Sox2為Atoh1的上游調控因子,通過直接互作協同調節Atoh1增強子,誘導耳蝸非感覺上皮產生異位新生毛細胞。在新生小鼠中大上皮嵴中同時過表達Atoh1和胰島素增強子結合蛋白1(insulin enhancer binding protein 1, Isl1)可有效增強毛細胞轉化效率[27]。新霉素損傷耳蝸毛細胞后,共轉染配對盒子基因2(paired box 2, Pax2)和Atoh1能誘導支持細胞原位增殖和轉分化為再生毛細胞[28]。在成年小鼠耳蝸支持細胞中同時誘導Atoh1和Ikaros家族鋅指轉錄因子2(Ikaros family zinc finger 2, Ikzf2兩個外毛細胞發育關鍵轉錄因子,可將支持細胞轉化為Prestin特異性表達的外毛細胞樣細胞[29]。若將Atoh1及其輔因子T細胞因子3(T-cell factor3, Tcf3)、Gata3或ETS變異體4(ETS Translocation Variant 4,Etv4)、Mycn原癌基因(MYCN proto-oncogene, Nmyc)、E26轉錄因子2 (E-twenty six 2, Ets2)聯合傳遞到耳蝸上皮也可誘導耳蝸非感覺細胞再生毛細胞[30-31]。關于Gfi1、Pou4f3和Atoh1組合表達(GPA)的研究顯示,GPA可有效誘導雞胚耳上皮細胞直接重編程為毛細胞樣細胞[32],Chen[33]等再次驗證了GPA是功能性毛細胞再生必不可少的基因,且GPA因子與另一轉錄因子Six1可在體外將小鼠胚胎成纖維細胞和尾尖成纖維細胞直接重編程為毛細胞樣細胞[34]。上述研究結果表明:結合不同的轉錄因子對增強Atoh1的毛細胞重編程效率是必要的。其中,Gfi1和Pou4f3與Atoh1互作促毛細胞再生效率、功能建立和存活的效果尤為顯著。

Figure 1. Interactions of atonal bHLH transcription factor 1 (Atoh1) with other hair cell development-related factors and their signaling pathways. Pou4f3: POU domain class 4 transcription factor 3; Gfi1: growth factor independent-1; E2f1: E2F transcription factor 1; Hic1: hypermethylated in cancer 1; Barhl1: BarH-like homeobox 1; Zic1: Zic family member 1; Six1:sineoculis homeobox homolog 1; Sox2: sex-determining region Y-box 2; Neurog1: neurogenin 1; Neurod1: neurogenic differentiation factor1; Id1-3: inhibitor of differentiation 1-3; Cdk-2: cyclin-dependent kinase-2; Hey1: hairy/enhancer-of-split related with YRPW motif 1; Hey2: hairy/enhancer-of-split related with YRPW motif 2; Pias3: protein inhibitor of activated signal transducer and activator of transcription 3; Stat3: signal transducer and activator of transcription 3; Gata3: GATA binding protein3; p27kip1: cyclin-dependent kinase inhibitor 1. → refers to activation; ⊥ refers to inhibition; ├┤ refers to antagonism; ┄ refers to predicted.
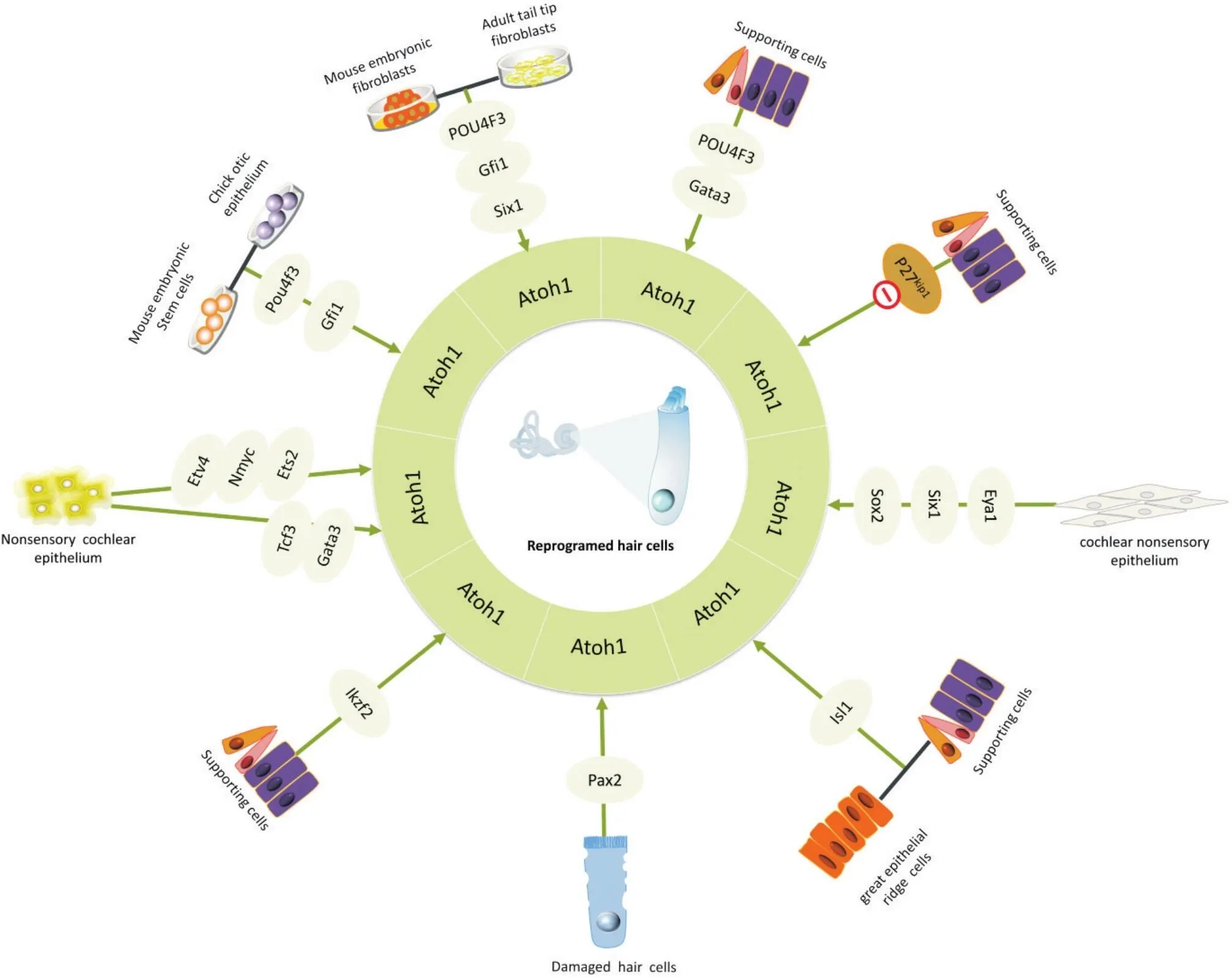
Figure 2. Hair cell reprogramming strategies of atonal bHLH transcription factor 1 (Atoh1) in combination with additional transcription factors. Pou4f3: POU domain class 4 transcription factor 3; Gata3: GATA binding protein 3; p27kip1: cyclin-dependent kinase inhibitor 1; Eya1: eyes absent 1; Six1: sineoculis homeobox homolog 1; Sox2: sex-determining region Y-box 2; Isl1: insulin enhancer binding protein 1; Pax2: paired box 2; Ikzf2: Ikaros family zinc finger 2; Tcf3: transcription factor 3; Etv4: ETS translocation variant 4; Nmyc: MYCN proto-oncogene, bHLH transcription factor; Ets2:E-twenty six 2; Gfi1: growth factor independent-1. Red refers to knockout.
2 Pou4f3和Gfi1在內耳中的結構與功能
2.1Pou4f3在內耳中的功能及其效應基因(亦稱或)基因是調節神經內分泌途徑的第Ⅳ類POU結構域轉錄因子,含有2個外顯子,編碼338個氨基酸,位于人類染色體5q31-q33。突變會導致常染色體顯性非綜合征型耳聾DFNA15。敲除小鼠在內耳分化初始階段無形態異常,毛細胞表達、、等特異性標志基因,隨后部分毛細胞表現異常,未能形成正常毛束結構并逐漸發生凋亡,許多螺旋和前庭神經節細胞丟失,出現全聾和前庭系統重度受損的癥狀[35]。Weiss等[36]通過免疫定位明確的正常產物定位于細胞核,突變蛋白定位于細胞質和細胞核,的細胞質定位可能阻止其作為轉錄調節因子發揮作用。馬登濱[37]在透射電鏡下觀察到基因突變小鼠內毛細胞出現線粒體空化現象。線粒體作為細胞能量供應器,其形態結構異常會使毛細胞能量供應受損以致功能障礙,提示毛細胞功能障礙可能與自身超微結構發生變化有關[38]。上述研究為基因突變致小鼠耳聾提供了可能的細胞學機制。
目前參與毛細胞分化和功能維持的分子途徑有待深入研究,其下游靶基因的鑒定將有助于闡明突變導致人類聽力損失的分子機制。已知的直接靶點有腦源性神經營養因子(neurotrophins brain derived neurotrophic factor,Bdnf)、神經營養素3(neurotrophin-3,Nt-3)、細胞質激活增殖相關蛋白1(cytoplasmic activation and proliferation-associated protein-1,Caprin-1)和核受體2家族2(nuclear receptor subfamily 2 group fmember 2,Nr2f2)[39-40],以及下游靶標LIM同源盒3(LIM-homeodomain 3,Lhx3)和Gfi1[39-42]。神經生長因子Bdnf和Nt-3在前庭毛細胞及支持細胞中均有表達,Bdnf主要在毛細胞中表達,Nt-3則主要表達于支持細胞[43-47]。Pou4f3敲除鼠的Bdnf和Nt-3表達量均有下降,表現出耳蝸神經支配缺陷和感覺神經元丟失[40],間接影響毛細胞發育。Caprin-1在毛細胞和支持細胞中表達,其5′-端側翼序列包含Pou4f3結合位點,介導Pou4f3對Caprin-1表達的抑制作用。氨基糖苷類藥物損傷耳蝸后,內耳毛細胞被誘導生成含有Caprin-1的應激顆粒,參與毛細胞的應激反應[48]。Nr2f2是一種類固醇/甲狀腺激素核受體,其5′-側翼區包含Pou4f3的兩個結合位點,參與耳蝸基底膜的延伸發育。Tornari等[49]采用消減雜交法將鑒定為Pou4f3的靶基因,Pou4f3突變致兩個結合位點活性減弱,Nr2f2表達相應下調,繼而影響內耳功能。Lhx3屬LIM 同源域轉錄因子家族的一員,在小鼠E16特異性表達于內耳毛細胞中,其在耳蝸中受Pou4f3調控,但在前庭毛細胞中不受調控[50]。此外Gfi1作為Pou4f3下游靶基因,也是毛細胞發育和存活的重要轉錄因子(圖3)。闡明在毛細胞中的作用對判斷其能否成為毛細胞再生直接重編程的候選基因至關重要。
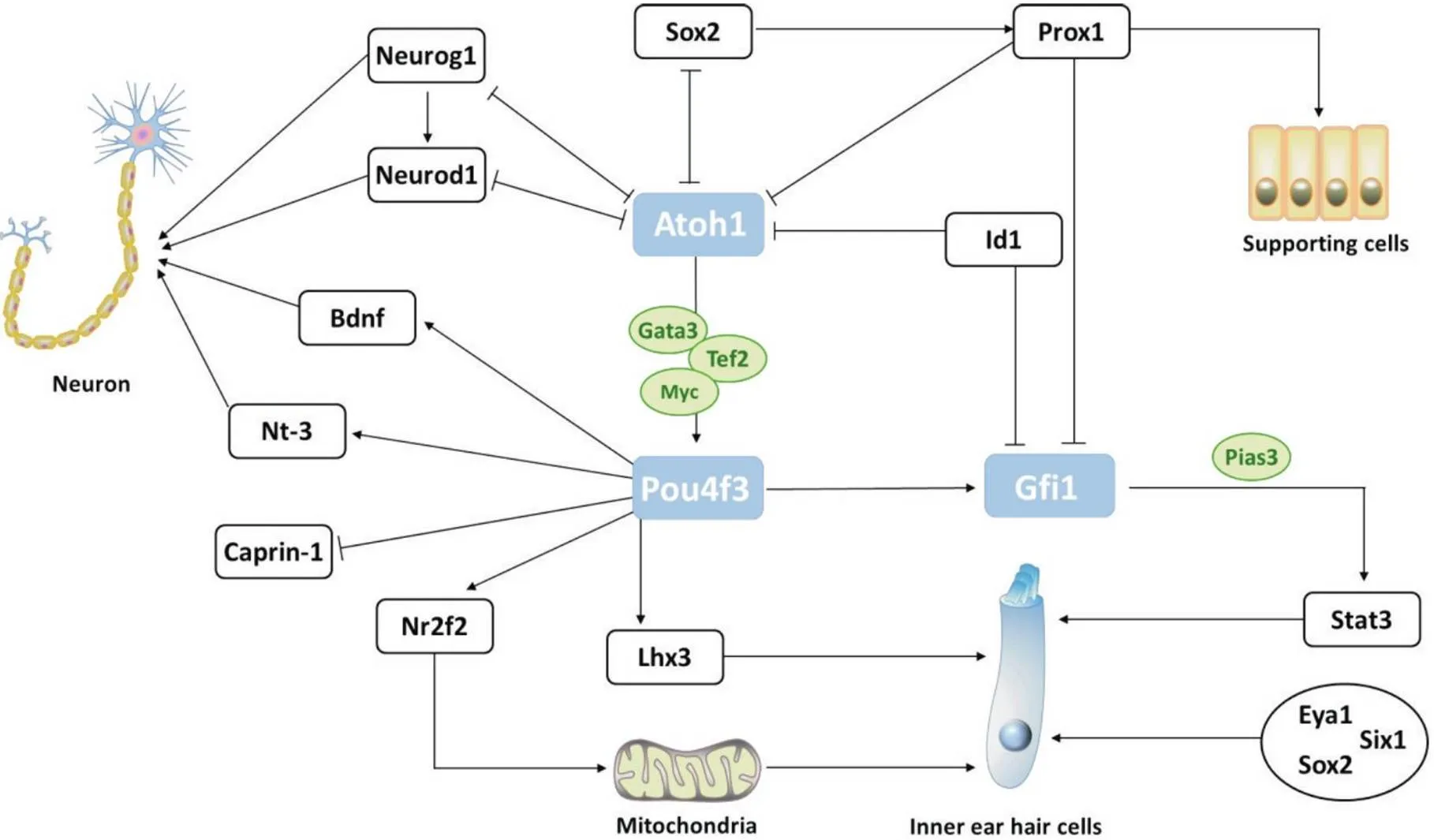
Figure 3. POU domain class 4 transcription factor 3 (Pou4f3) downstream target genes and the mechanistic relationship between atonal bHLH transcription factor 1 (Atoh1)-Pou4f3-growth factorindependent-1 (Gfi1). Sox2:sex-determining region Y-box 2; Neurog1: neurogenin 1; Neurod1: neurogenic differentiation factor1; Bdnf: brain-derived neurotrophic factor; Nt-3: neurotrophin-3; Caprin-1: cytoplasmic activation and proliferation-associated protein-1; Nr2f2: nuclear receptor subfamily 2 group F member 2; Lhx3: LIM-homeodomain 3; Stat3: signal transducer and activator of transcription 3; Pias3: protein inhibitor of activated signal transducer and activator of transcription 3; Prox1: Prospero-related homeobox protein 1; Id1: inhibitor of differentiation 1; Gata3: GATA binding protein 3; Tef2: transcription factor 3; Myc: MYC proto-oncogene, bHLH transcription factor; Six1: sineoculis homeobox homolog 1; Eya1: eyes absent 1. → refers to activation; ⊥ refers to inhibition ; ├┤refers to antagonism.
2.2在內耳毛細胞中的功能的C末端含有由六個C2H2鋅指結構組成的DNA結合結構域,N末端含有20個氨基酸的SNAG阻遏物結構域和一個保守的核定位信號。小鼠內耳發育早期敲除不會嚴重破壞毛細胞形態結構,但隨著分化進行,毛細胞缺陷會逐漸明顯:在E16.5時毛細胞紊亂,到E18.5時外毛細胞、內毛細胞先后由基底至頂端呈凋亡跡象,至P14時所有內耳毛細胞均消失。由此可見,缺失會導致毛細胞發育異常和程序性死亡。Hertzano等[42]鑒定轉錄激活蛋白3(signal transducer and activator of transcription 3,Stat3)為的下游效應蛋白,定位于耳蝸感覺上皮的外毛細胞中,參與調控細胞的增殖、分化和凋亡等過程。與轉錄激活子3的蛋白抑制劑(protein inhibitor of activated signal transducer and activator of transcription 3, Pias3)相互作用可增強信號[51],推測Gfi1在內耳毛細胞中的作用機制是通過與相互作用以促進毛細胞分化(圖3)。Matern等[52]使用Gfi1小鼠模型對缺失的毛細胞翻譯組進行分析,顯示神經元相關基因(如)上調,而毛細胞功能相關基因表達量顯著降低。提示作為干細胞分化為毛細胞命運的特異性基因,可能通過抑制神經元為主的非毛細胞基因的表達,并激活毛細胞特異基因的表達。
總之,、和在毛細胞分化、成熟和聽覺功能建立中均不可或缺,可作為毛細胞再生直接重編程的候選治療基因,分析三者在毛細胞功能重塑中的互作機制以便更好地指導并實現毛細胞再生。
3 Atoh1-Pou4f3-Gfi1的互作機制與直接重編程策略
3.1Atoh1與Pou4f3互作Pou4f3和Gfi1均為Atoh1的下游靶點,具有相似的表達梯度。但與缺失不同的是,缺失導致的表型缺陷更嚴重:缺失不僅導致耳蝸及前庭毛細胞凋亡,且細胞凋亡在胚胎期便已有跡象,靜纖毛束形態缺陷也比基因敲除小鼠更嚴重[53]。Yu等[54]指出在E15.5,和共表達于耳蝸底部四排毛細胞中,而分化程度較低的頂端只檢測到表達。Ohyama等[55]使用轉基因小鼠在E8.5敲除后,在E15.5耳蝸感覺上皮中未檢測到Pou4f3蛋白。可見,在毛細胞分化過程中,Pou4f3位于Atoh1下游且受Atoh1調控。Ikeda等[31]報道可與、和協同調控基因和毛細胞表型(圖3)。此外,也可驅動與異染色質結合而解壓縮DNA,使能激活染色質中的毛細胞分化基因網絡以促進毛細胞分化程序[54]。
3.2Atoh1與Gfi1互作作為一種調節細胞周期的原癌基因,具有促進細胞增殖和抑制細胞凋亡的功能。García-A?overos等[56]闡明T-BOX轉錄因子2(T-BOX transcription factor 2,)是內毛細胞特異性分化及整個發育過程所必需的基因,而Atoh1和Gfi1的存在是Tbx2行使內毛細胞命運決策因子的前提條件。提示Atoh1和Gfi1是后續獲取大量毛細胞及實現毛細胞功能修復必不可少的基因。使用成年小鼠毛細胞損傷模型的體內研究表明,Corti器中腺病毒介導的Atoh1和Gfi1過表達會使大量支持細胞轉分化為毛細胞樣細胞,再生效率顯著高于單獨過表達Atoh1[57]。Wallis等[58]證明Gfi1敲除鼠的Atoh1表達量并無明顯異常,推測Atoh1的表達可能不受Gfi1影響。Atoh1突變小鼠Gfi1的mRNA量雖無明顯變化,但聽覺上皮中Gfi1蛋白表達量卻明顯下降甚至消失,說明Atoh1為Gfi1的上游調控基因且對Gfi1的調控位于翻譯水平。Prospero相關同源異形盒蛋白1(prospero related homeobox protein 1, Prox1)為Sox2的下游靶基因,不僅可負調控Gfi1且介導Sox2對Atoh1的抑制作用[59]。此外,Gfi1可通過抑制Id1以保持Atoh1的表達水平[42],如圖3所示。
3.3Pou4f3與Gfi1互作在耳蝸發育過程中,蛋白在胚胎期E 13.5~14.5之間的內耳祖細胞基部至頂部表達,驅使內耳祖細胞向毛細胞分化,并在E14.5至E16激活Pou4f3的表達,Pou4f3隨后在E16.5激活Gfi1的表達。Pou4f3是毛細胞分化過程中Atoh1的直接靶基因,Gfi1則為Pou4f3的靶基因[58]。研究發現,在Pou4f3–/–或Pou4f3ddl/ddl小鼠中,Pou4f3的缺失將導致Gfi1的表達水平在統計學上顯著降低,與Pou4f3的表達量存在正相關性[42]。馮曉等[60]曾報道Gfi1表達量在野生型、雜合型以及完全缺失Pou4f3的斑馬魚神經組織中有明顯的量效差別,二者與神經組織發育有關。總之,Pou4f3對Gfi1的表達具有調控作用,我們推測:胚胎后期Pou4f3正向調控Gfi1之后,Gfi1可能通過Pias3/Stat3級聯作用或負調控Id1以維持Atoh1的表達水平,從而促進毛細胞分化和成熟。
3.4Atoh1-Pou4f3-Gfi1直接重編程策略重編程轉錄因子通過改變起始細胞轉錄組和表觀遺傳景觀,從而達到激活某些特定基因的目的,以促進特定細胞類型轉錄程序。Atoh1通過與Gfi1和Pou4f3的聯合調控可將神經元譜系轉變為內耳毛細胞譜系。僅過表達Atoh1會促使神經元分化,Pou4f3通過激活起始細胞原來封閉的染色質區域,使Atoh1能夠參與并激活驅動分化為毛細胞的基因程序,但Atoh1和Pou4f3不足以促進毛細胞分化。Gfi1是Atoh1誘導毛細胞譜系轉換的關鍵,通過抑制Atoh1對毛細胞分化拮抗基因的誘導,同時增強Atoh1促毛細胞特異性基因表達的能力,進而啟動毛細胞轉錄程序[54]。Chen等[33]為實現毛細胞功能重塑,構建了支持細胞中同時過表達的轉基因小鼠,Lgr5-GPA-tdTomato小鼠中Myo7a+/tdTomato+細胞數量顯著高于Lgr5-Atoh1-tdTomato小鼠,新生毛細胞具有電生理活性,證明Pou4f3和Gfi1在Atoh1介導的支持細胞直接轉分化為毛細胞過程中起正向調節作用。綜上所述,GPA聯合調控能夠從根本上改變Atoh1轉錄程序以促毛細胞分化,提高細胞直接重編程效率,同時促進新生毛細胞亞型分化和電生理功能成熟。Atoh1、Gfi1和Pou4f3是促進毛細胞分化的特異性轉錄因子,Atoh1-Pou4f3-Gfi1(GPA)的協同調控是當前毛細胞再生直接重編程的可行方案。
5 總結和展望
內耳毛細胞受損導致的感音神經性耳聾嚴重影響著患者的生活質量和社會交流能力,其再生性功能修復的問題一直備受關注。近年來,Atoh1-Pou4f3-Gfi1直接重編程技術通過病毒攜帶毛細胞發育所需關鍵轉錄因子編碼基因感染支持細胞或成纖維細胞使其直接轉換為毛細胞,為后續臨床治療耳聾提供了一種新思路。但三者在毛細胞分化和功能重塑中的作用機制及上下游靶基因卻鮮有報道。因此,本文總結了Atoh1、Pou4f3、Gfi1及其相關的上下游基因在毛細胞發育、成熟、聽覺功能建立中的作用及互作機制,并提出Atoh1-Pou4f3-Gfi1直接重編程技術是未來治療聽力損失的可行性方案。當前大部分直接重編程研究主要在體外或模式動物中進行的,后續可在人成纖維細胞中過表達獲取內耳毛細胞,并移植到人類內耳類器官損傷模型,通過分析其功能修復能力,為耳聾治療方法的實驗研究和臨床應用提供參考。
[1] Wang H, Yang Y, Liu J, et al. Direct cell reprogramming: approaches, mechanisms and progress[J]. Nat Rev Mol Cell Biol, 2021, 22(6):410-424.
[2]任麗偉,楊曉菲,李富榮. 細胞直接重編程——治療糖尿病的新技術[J]. 中國病理生理雜志, 2014, 30(12):2284-2288.
Ren LW, Yang XF, Li FR. Cell direct reprogramming: a new technique for treating diabetes[J]. Chin J Pathophysiol, 2014, 30(12):2284-2288.
[3] Kong L, Xin Y, Chi F, et al. Developmental and functional hair cell-like cells induced by Atoh1 overexpression in the adult mammalian cochlea[J]. Neural Plast, 2020, 2020:8885813.
[4] Iyer AA, Groves AK. Transcription factor reprogramming in the inner ear: turning on cell fate switches to regenerate sensory hair cells[J]. Front Cell Neurosci, 2021, 15:660748.
[5] Schimmang T. Transcription factors that control inner ear development and their potential for transdifferentiation and reprogramming[J]. Hear Res, 2013, 297:84-90.
[6] Liu Z, Dearman JA, Cox BC, et al. Age-dependentconversion of mouse cochlear pillar and deiters' cells to immature hair cells by Atoh1 ectopic expression[J]. J Neurosci, 2012, 32(19):6600-6610.
[7] Duran Alonso MB, Lopez Hernandez I, de la Fuente MA, et al. Transcription factor induced conversion of human fibroblasts towards the hair cell lineage[J]. PLoS One, 2018, 13(7):e0200210.
[8] Maricich SM, Wellnitz SA, Nelson AM, et al. Merkel cells are essential for light-touch responses[J]. Science, 2009, 324(5934):1580-1582.
[9] Westerman BA, Breuer RH, Poutsma A, et al. Basic helix-loop-helix transcription factor profiling of lung tumors shows aberrant expression of the proneural gene atonal homolog 1 (ATOH1, HATH1, MATH1) in neuroendocrine tumors[J]. Int J Biol Markers, 2007, 22(2):114-123.
[10] Bossuyt W, Kazanjian A, De Geest N, et al. Atonal homolog 1 is a tumor suppressor gene[J]. PLoS Biol, 2009, 7(2):e39.
[11] Flora A, Klisch TJ, Schuster G, et al. Deletion of Atoh1 disrupts sonic hedgehog signaling in the developing cerebellum and prevents medulloblastoma[J]. Science, 2009, 326(5958):1424-1427.
[12] Lumpkin EA, Collisson T, Parab P, et al. Math1-driven GFP expression in the developing nervous system of transgenic mice[J]. Gene Expr Patterns, 2003, 3(4):389-395.
[13] Stojanova ZP, Kwan T, Segil N. Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea[J]. Development, 2015, 142(20):3529-3536.
[14] Albu S, Muresanu DF. Vestibular regeneration--experimental models and clinical implications[J]. J Cell Mol Med, 2012, 16(9):1970-1977.
[15] Cai T, Seymour ML, Zhang H, et al. Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti[J]. J Neurosci, 2013, 33(24):10110-10122.
[16] Walters BJ, Coak E, Dearman J, et al.interplay between p27Kip1, GATA3, ATOH1, and POU4F3 converts non-sensory cells to hair cells in adult mice[J]. Cell Rep, 2017, 19(2):307-320.
[17] Cheng YF. Atoh1 regulation in the cochlea: more than just transcription[J]. J Zhejiang Univ Sci B, 2019, 20(2):146-155.
[18] Gomez-Dorado M, Daudet N, Gale JE, et al. Differential regulation of mammalian and avian ATOH1 by E2F1 and its implication for hair cell regeneration in the inner ear[J]. Sci Rep, 2021, 11(1):19368.
[19] Ebert PJ, Timmer JR, Nakada Y, et al. Zic1 represses Math1 expression via interactions with the Math1 enhancer and modulation of Math1 autoregulation[J]. Development, 2003, 130(9):1949-1959.
[20] Briggs KJ, Eberhart CG, Watkins DN. Just say no to ATOH: how HIC1 methylation might predispose medulloblastoma to lineage addiction[J]. Cancer Res, 2008, 68(21):8654-8656.
[21] Jahan I, Pan N, Kersigo J, et al. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea[J]. PLoS One, 2010, 5(7):e11661.
[22] Zhang T, Xu J, Maire P,et al. Six1 is essential for differentiation and patterning of the mammalian auditory sensory epithelium[J]. PLoS Genet, 2017, 13(9):e1006967.
[23] Benito-Gonzalez A, Doetzlhofer A. Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of hedgehog signaling[J]. J Neurosci, 2014, 34(38):12865-12876.
[24] Benezra R, Davis RL, Lockshon D, et al. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins[J]. Cell, 1990, 61(1):49-59.
[25] Kamaid A, Neves J, Giraldez F. Id gene regulation and function in the prosensory domains of the chicken inner ear: a link between Bmp signaling and Atoh1[J]. J Neurosci, 2010, 30(34):11426-11434
[26] Ahmed M, Wong EY, Sun J, et al. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2[J]. Dev Cell, 2012, 22(2):377-390.
[27] Yamashita T, Zheng F, Finkelstein D, et al. High-resolution transcriptional dissection of in vivo Atoh1-mediated hair cell conversion in mature cochleae identifies Isl1 as a co-reprogramming factor[J]. PLoS Genet, 2018, 14(7):e1007552.
[28] Chen Y, Yu H, Zhang Y, et al. Cotransfection of Pax2 and Math1 promote in situ cochlear hair cell regeneration after neomycin insult[J]. Sci Rep, 2013, 3:2996.
[29] Sun S, Li S, Luo Z, et al. Dual expression of Atoh1 and Ikzf2 promotes transformation of adult cochlear supporting cells into outer hair cells[J]. Elife, 2021, 10:e66547.
[30] Masuda M, Pak K, Chavez E, et al. Tfe2 and Gata3 enhance induction of Pou4f3 and myosin VIIa positive cells in nonsensory cochlear epithelium by Atoh1[J]. Dev Biol, 2012, 372(1):68-80.
[31] Ikeda R, Pak K, Chavez E, et al. Transcription factors with conserved binding sites near Atoh1 on the Pou4f3 gene enhance the induction of cochlear hair cells[J]. Mol Neurobiol, 2015, 51(2):672-684.
[32] Costa A, Sanchez-Guardado L, Juniat S, et al. Generation of sensory hair cells by genetic programming with a combination of transcription factors[J]. Development, 2015, 142(11):1948-1959.
[33] Chen Y, Gu Y, Li Y, et al. Generation of mature and functional hair cells by co-expression of Gfi1, Pou4f3, and Atoh1 in the postnatal mouse cochlea[J]. Cell Rep, 2021, 35(3):109016.
[34] Menendez L, Trecek T, Gopalakrishnan S, et al. Generation of inner ear hair cells by direct lineage conversion of primary somatic cells[J]. Elife, 2020, 9:55249.
[35] Xiang M, Gan L, Li D, et al. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development[J]. Proc Natl Acad Sci U S A, 1997, 94(17):9445-9450.
[36] Weiss S, Gottfried I, Mayrose I, et al. The DFNA15 deafness mutation affects Pou4f3 protein stability, localization, and transcriptional activity[J]. Mol Cell Biol, 2003, 23(22):7957-7964.
[37] 馬登濱. Pou4f3基因突變致小鼠耳聾的聽力學機制研究[D]. 南京:南京醫科大學, 2013:30-32.
Ma DB. The studies of hearing mechanism in Pou4f3 mutant mice[D]. Nanjing: Nanjing Medical University, 2013.
[38] 熊燕,張梅,陳菲,等. 線粒體功能障礙與心血管疾病[J]. 中國病理生理雜志, 2013, 29(2):364-370.
Xiong Y, Zhang M, Chen F, et al. Roles of mitochondrial dysfunction in cardiovascular diseases[J]. Chin J Pathophysiol, 2013, 29(2):364-370.
[39] Towers ER, Kelly JJ, Sud R, et al. Caprin-1 is a target of the deafness gene Pou4f3 and is recruited to stress granules in cochlear hair cells in response to ototoxic damage[J]. J Cell Sci, 2011, 124(Pt 7):1145-1155.
[40] Clough RL, Sud R, Davis-Silberman N, et al. Brn-3c (POU4F3) regulates BDNF and NT-3 promoter activity[J]. Biochem Biophys Res Commun, 2004, 324(1):372-381.
[41] Hertzano R, Dror AA, Montcouquiol M, et al. Lhx3, a LIM domain transcription factor, is regulated by Pou4f3 in the auditory but not in the vestibular system[J]. Eur J Neurosci, 2007, 25(4):999-1005.
[42] Hertzano R, Montcouquiol M, Rashi-Elkeles S, et al. Transcription profiling of inner ears fromPou4f3/ddlidentifies Gfi1 as a target of the Pou4f3 deafness gene[J]. Hum Mol Genet, 2004, 13(18):2143-2153.
[43] Fritzsch B, Silos-Santiago I, Bianchi LM, et al. The role of neurotrophic factors in regulating the development of inner ear innervation[J]. Trends Neurosci, 1997, 20(4):159-164.
[44] Ernfors P, Lee KF, Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits[J]. Nature, 1994, 368(6467):147-150.
[45] Ernfors P, Lee KF, Kucera J, et al. Lack of neurotrophin-3 leads to deficiencies in the peripheral nervous system and loss of limb proprioceptive afferents[J]. Cell, 1994, 77(4):503-512.
[46] Agerman K, Hjerling-Leffler J, Blanchard MP, et al.gene replacement reveals multiple mechanisms for establishing neurotrophin specificity during sensory nervous system development[J]. Development, 2003, 130(8):1479-1491.
[47] Ernfors P, Van De Water T, Loring J, et al. Complementary roles of BDNF and NT-3 in vestibular and auditory development[J]. Neuron, 1995, 14(6):1153-1164.
[48] Solomon S, Xu Y, Wang B, et al. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs[J]. Mol Cell Biol, 2007, 27(6):2324-2342.
[49] Tornari C, Towers ER, Gale JE, et al. Regulation of the orphan nuclear receptor Nr2f2 by the DFNA15 deafness gene Pou4f3[J]. PLoS One, 2014, 9(11):e112247.
[50] Huang M, Sage C, Li H, et al. Diverse expression patterns of LIM-homeodomain transcription factors (LIM-HDs) in mammalian inner ear development[J]. Dev Dyn, 2008, 237(11):3305-3312.
[51] Rodel B, Tavassoli K, Karsunky H, et al. The zinc finger protein Gfi-1 can enhance Stat3 signaling by interacting with the Stat3 inhibitor Pias3[J]. Embo J, 2000, 19(21):5845-5855.
[52] Matern MS, Milon B, Lipford EL, et al. Gfi1 functions to repress neuronal gene expression in the developing inner ear hair cells[J]. Development, 2020, 147(17):dev186015.
[53] Xiang M, Gao WQ, Hasson T, et al. Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells[J]. Development, 1998, 125(20):3935-3946.
[54] Yu HV, Tao L, Llamas J, et al. Pou4f3 pioneer activity enables Atoh1 to drive diverse mechanoreceptor differentiation through a feed-forward epigenetic mechanism[J]. Proc Natl Acad Sci U S A, 2021, 118(29):e2105137118.
[55] Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome[J]. Genesis, 2004, 38(4):195-199.
[56] García-A?overos J, Clancy JC, Foo CZ, et al. Tbx2 is a master regulator of inner versus outer hair cell differentiation[J]. Nature, 2022, 605(7909):298-303.
[57] Lee S, Song JJ, Beyer LA, et al. Combinatorial Atoh1 and Gfi1 induction enhances hair cell regeneration in the adult cochlea[J]. Sci Rep, 2020, 10(1):21397.
[58] Wallis D, Hamblen M, Zhou Y, et al. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival[J]. Development, 2003, 130(1):221-232.
[59] Dabdoub A, Puligilla C, Jones JM, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea[J]. Proc Natl Acad Sci U S A, 2008, 105(47):18396-18401.
[60] 馮曉,齊麟,孟娟.斑馬魚耳聾基因Gfi與POU4f3的關聯性[J]. 河南大學學報(醫學版), 2014, 33(4):267-269.
Feng X, Qi L, Meng J. The relation between Gfi and Pou4f3 by zebrafish[J]. J Henan Univ (Med Sci), 2014, 33(4):267-269.
Advances in study ofgene synergizing with POU4F3 and Gfi1 to promote hair cell regeneration
He Cairong1, Yang Ying1, Huang Jinwei1, Chen Fengjiao2, Lu Ying3, Ding Jie1△
[1(),,,550025,;2,550002,;3,,,550025,]
Hair cell regeneration and functional remodeling is an effective method of treating sensorineural deafness. In recent years, direct reprogramming techniques have made substantial progress in the field of hair cell regeneration.is an essential gene that is involved in hair cell morphology and maturation. POU domain class 4 transcription factor 3 (Pou4f3)plays a crucial role in the development of all hair cells within the sensory epithelium.Growth factor indepondent-1 (Gfi1) is an important transcription factor that is required for hair cell development and survival. Studies have demonstrated that the synergistic regulation of Atoh1, Pou4f3 and Gfi1 promotes hair cell differentiation, as well as functional maturation,. However, the regulatory mechanisms of these three factors in the development, maturation, and establishment of the auditory function of hair cells are still to be further explored. The aim of this paper is to elucidate the function and mechanistic relationship between Atoh1, Pou4f3 and Gfi1 in the functional remodeling of hair cells, as well as to determine the feasibility of using the Atoh1-Pou4f3-Gfi1 direct reprogramming strategy in order to restore human hearing in the future.
hair cell regeneration; direct reprogramming; interoperability mechanisms; hearing loss
R363; R763.3
A
10.3969/j.issn.1000-4718.2023.09.016
1000-4718(2023)09-1666-09
2023-03-15
2023-06-19
國家自然科學基金資助項目(No. 32260163; No. 81960838);貴州省科技計劃項目(黔科合基礎?ZK[2021]一般108);貴州省高層次創新人才(黔科合平臺人才-GCC[2022]027-1)
Tel: 18085108128; E-mail: 47734092@qq.com
(責任編輯:余小慧,羅森)

