不同生長環境下水稻氮、磷、鉀利用相關性狀的QTL定位分析
楊樹明, 曾亞文, 王 荔, 杜 娟, 普曉英, 楊 濤
(1云南省農業科學院生物技術與種質資源研究所, 云南昆明 650205;2農業部西南農業基因資源與種質創制重點實驗室, 云南昆明 650223;3云南省農業生物技術重點實驗室, 云南昆明 650223; 4云南農業大學農學與生物技術學院, 云南昆明 650201)
不同生長環境下水稻氮、磷、鉀利用相關性狀的QTL定位分析
楊樹明1,2, 曾亞文1, 3*, 王 荔4*, 杜 娟1, 普曉英1, 楊 濤1
(1云南省農業科學院生物技術與種質資源研究所, 云南昆明 650205;2農業部西南農業基因資源與種質創制重點實驗室, 云南昆明 650223;3云南省農業生物技術重點實驗室, 云南昆明 650223; 4云南農業大學農學與生物技術學院, 云南昆明 650201)

水稻(OryzasativaL.); 近等基因系; 養分吸收; 數量性狀基因座; QTL多效性

1 材料與方法
1.1 作圖群體
以云南孕穗期強耐冷(2級)地方稻種麗江新團黑谷與十和田雜交、回交,并結合耐冷鑒定培育的包含105個株系的孕穗期耐冷性近等基因系(十和田4//麗江新團黑谷/十和田)BC4F8群體[23-24]。
1.2 試驗設計

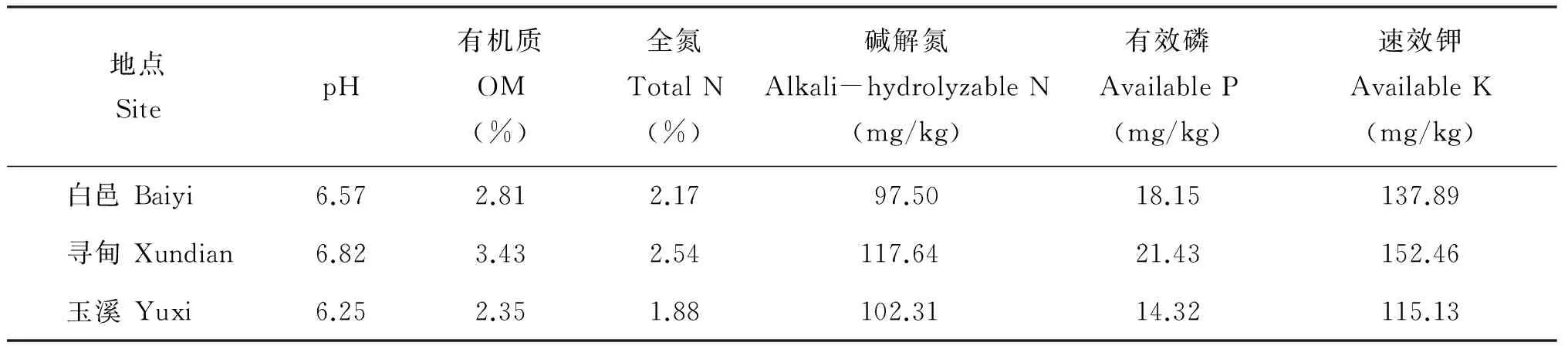
表1 不同生態點土壤理化性質Table 1 Physical and chemical properties of soils from three eco-sites
1.3 測定指標與方法
1.3.1 產量測定 成熟期,除邊行株外,各小區隨機選取代表性植株10兜齊地收割、風干,分成稻谷和稻草兩部分,測定單株籽粒干重、單株稻草干重和干物質總量,并留作氮、磷、鉀測定樣。每個家系以該10株的平均值為統計單元進行QTL分析。
1.3.2 葉片硝態氮含量及硝酸還原酶活性測定 孕穗至抽穗期,取每個家系和親本的劍葉裝入塑料袋封口,立即放入冰盒,保存于-80℃超低溫冰箱中。硝態氮含量和硝酸還原酶活性(NR)測定參照李合生等[25]的方法進行。
1.3.3 植株氮、磷、鉀含量測定 將上述測定產量時備留的稻谷和稻草樣品,分別用不銹鋼碾槽和微型植物粉碎機粉碎并過0.3mm篩,獲得的樣品用濃H2SO4-H2O2消煮,測定氮、磷和鉀含量。氮采用凱氏定氮法、磷為礬鉬黃比色法、鉀為火焰光度計法[26]。每個樣品重復測定2次,取其平均值為性狀表型值。
1.3.4 氮、磷和鉀吸收利用效率有關參數計算 3種養分的吸收利用效率有關參數計算參照文獻[27]進行。磷、鉀的計算公式與氮素相同,以氮為例,計算如下:
氮總吸收量(Total N accumulation,TNA)=稻谷產量×稻谷含氮量+稻草產量×稻草含氮量;
每100 kg籽粒需氮量(N absorption of 100-kg seeds)= 氮總吸收量/稻谷產量×100;
氮素干物質生產效率(N dry matter production efficiency,NDMPE)=干物質積累量/氮總吸收量;
氮素稻谷生產效率(N grain production efficiency,NGPE)=稻谷產量/氮總吸收量;
氮素收獲指數(N harvest index,NHI)=稻谷氮吸收量/氮總吸收量×100%。
1.4 基因型鑒定
1.4.1 田間取樣與DNA提取 在水稻分蘗期,對親本及BC4F8群體每個株系選1個單株(掛牌),取葉片上半部(5 g左右),按CTAB法[28]提取水稻全基因組DNA。
1.4.2 SSR標記檢測 按Temnykh等[29]方法進行SSR檢測。將模板DNA濃度調整到25 ng/μL,擴增反應體系為10 μL,含Taq酶0.2 μL(5 U/μL),Primer 1 0.5 μL,Primer 2 0.5 μL,10×PCR Buffer(含MgCl2) 1.5 μL,dNTPs 0.4 μL,DNA 2.0 μL,ddH2O 4.9 μL。PCR擴增程序為:94℃預變性5 min,94℃變性30 s,55℃退火40 s,72℃延伸45 s,35個循環后再72℃延伸5 min。擴增產物采用8%聚丙烯酰胺凝膠電泳分離,銀染檢測。
1.5 連鎖圖譜構建與QTL分析
應用QTL IciMapping V3.2軟件進行試驗數據的統計并構建遺傳連鎖圖譜進行QTL分析,共180個SSR標記,比較均勻分布于12個連鎖群。用MapDraw V2.1軟件繪圖,按Kosambi函數計算遺傳距離,構建的連鎖圖譜總共覆蓋水稻基因組約1820.6 cM,標記間平均距離為15.67 cM。參照王建康[30]的完備區間作圖法(Inclusive Composite Interval Mapping, ICIM),即ICIM-ADD,以P<0.005和LOD值>3.0為閾值來判斷QTL的存在。QTL的命名原則遵循McCouch[31]方法。
2 結果與分析
2.1 低溫脅迫和正常溫度下親本和NILs群體干物質量及氮素吸收利用相關性狀的表型分析
由表2可知,‘麗江新團黑谷’在單株籽粒干重、稻草干重、干物質總量、硝酸還原酶、硝態氮含量、稻谷和稻草含氮量、N總吸收量、每100 kg籽粒需氮量上為高值親本;而‘十和田’在氮素干物質生產效率、氮素稻谷生產效率和氮素收獲指數上均高于‘麗江新團黑谷’。BC4F8各性狀的平均值基本介于雙親之間,而白邑、尋甸的單株籽粒干重、硝酸還原酶活性、硝態氮含量、稻谷含氮量、氮素稻谷生產效率和氮素收獲指數低于玉溪,說明低溫環境對干物質積累及氮吸收利用影響較大。從變異范圍看,各性狀均出現一定數量的超親類型,而同一性狀的極值則低溫脅迫的變異度大于正常條件。正態分布檢驗表明,偏度除尋甸的硝酸還原酶活性、硝態氮含量以及玉溪的氮素收獲指數,而峰度除尋甸的硝酸還原酶活性、硝態氮含量、每100 kg籽粒需氮量以及玉溪硝酸還原酶活性、稻谷氮含量、稻草氮含量和氮素收獲指數絕對值略大于1外,其他性狀偏度和峰度絕對值都小于1,說明多數性狀呈類似正態的連續分布,適合QTL定位。
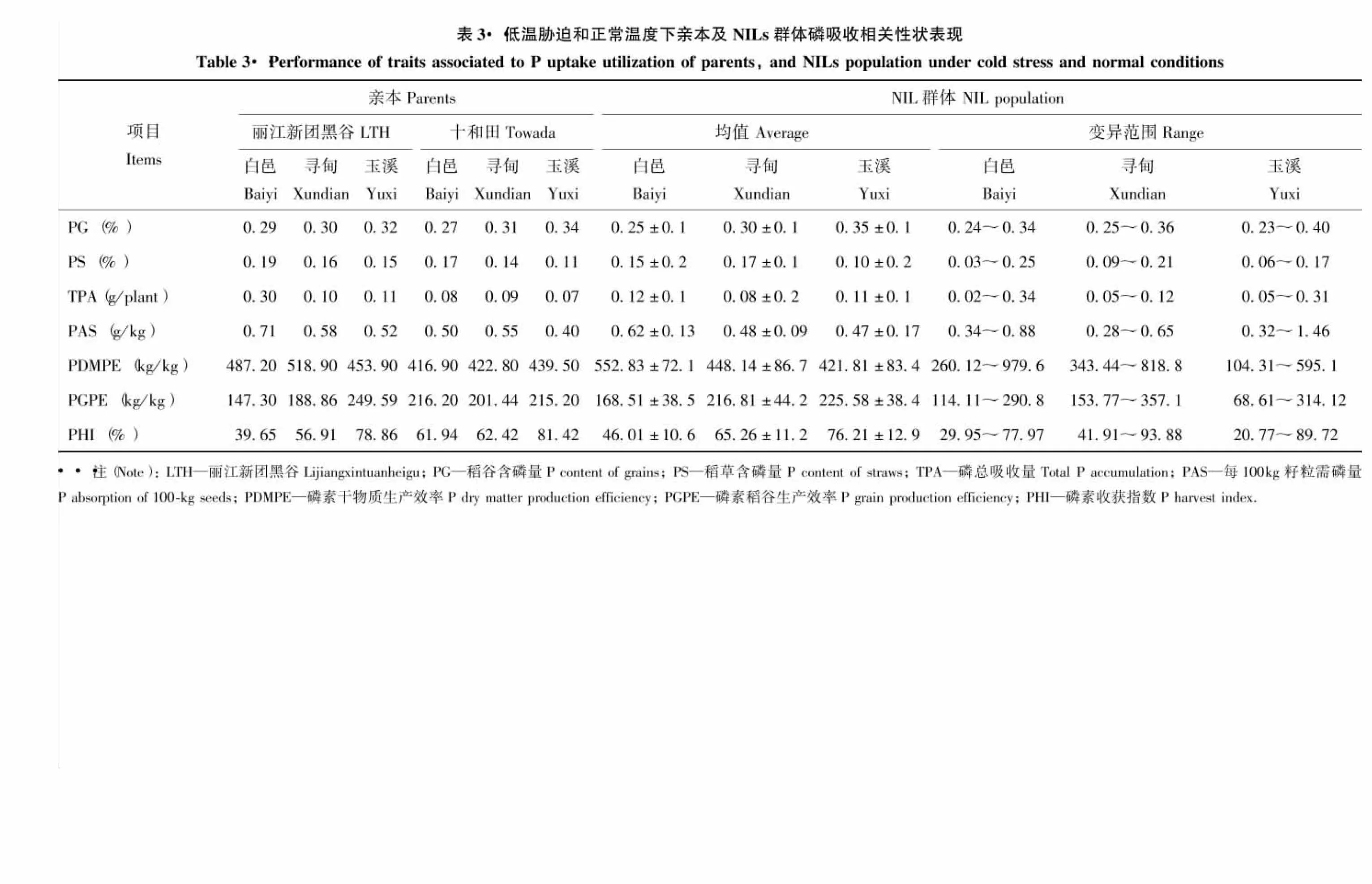
2.2 低溫脅迫和正常溫度下親本及NILs群體的磷素吸收利用相關性狀的表型分析
由表3可知,‘麗江新團黑谷’在稻草含磷量、磷總吸收量、每100 kg籽粒需磷量和磷素干物質生產效率上均明顯高于‘十和田’。BC4F8絕大多數指標的均值介于雙親之間,兩種低溫環境下的稻谷含磷量、每100 kg籽粒需磷量、磷素稻谷生產效率和磷素收獲指數均低于正常環境。從各性狀分布范圍看,同一性狀的極值呈現低溫脅迫變異更大的趨勢。偏度和峰度分析表明,偏度除白邑的稻谷磷含量、磷總吸收量和玉溪的磷總吸收量、每100 kg籽粒需磷量、磷素干物質生產效率、磷素稻谷生產效率、磷素收獲指數,而峰度除白邑的稻谷磷含量、磷總吸收量和尋甸的磷素干物質生產效率,以及玉溪的磷總吸收量、每100 kg籽粒需磷量、磷素干物質生產效率、磷素稻谷生產效率、磷素收獲指數絕對值略大于1外,其他性狀偏度和峰度的絕對值都小于1,說明多數性狀表型呈正態分布。
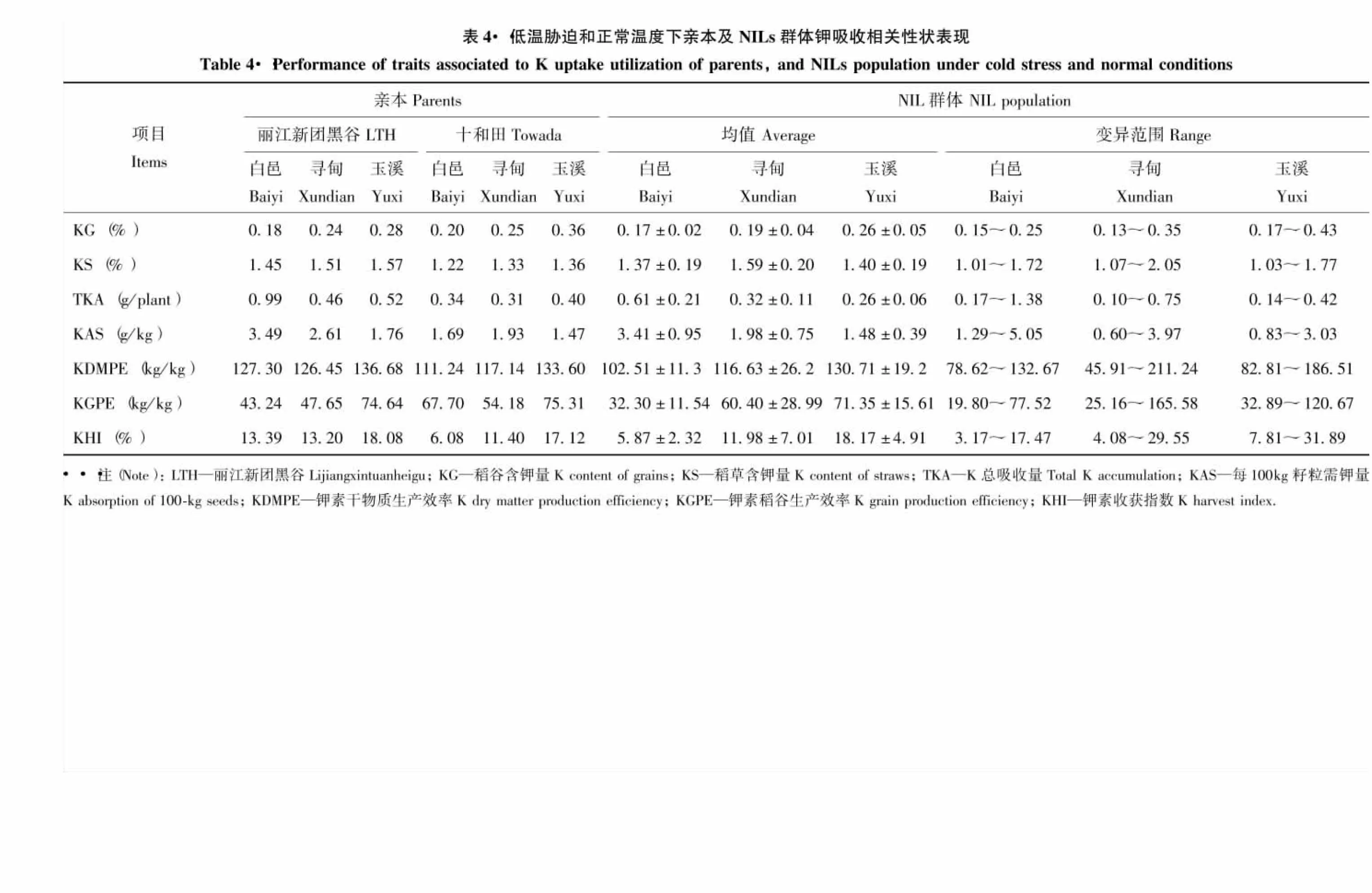
2.3 低溫脅迫和正常溫度下親本及NILs群體的鉀素吸收利用相關性狀的表型分析
由表4可知,水稻吸收鉀主要集中在稻草中。在兩種冷害脅迫下,稻草鉀含量、鉀素干物質生產效率、鉀素稻谷生產效率、鉀素收獲指數降低,其他指標則明顯增加。雙親的7個性狀差異明顯,其中‘麗江新團黑谷’的稻草含鉀量、鉀總吸收量、每100 kg籽粒需鉀量、鉀素干物質生產效率和鉀素收獲指數均高于‘十和田’。從群體的表型均值看,絕大多數性狀均值介于雙親之間。從各性狀變異范圍看,同一性狀的極值在不同環境中差異較大,但均呈連續變異,并存在明顯的雙向超親分離。偏度除白邑稻谷鉀含量、鉀素稻谷生產效率、鉀素收獲指數,尋甸鉀素稻谷生產效率及玉溪每100 kg籽粒需鉀量,而峰度除白邑和尋甸的稻谷鉀含量、鉀總吸收量、鉀素稻谷生產效率,以及玉溪每100 kg籽粒需鉀量絕對值略大于1外,其他性狀偏度和峰度的絕對值都小于1,說明多數性狀表型呈正態分布。
2.4 氮、磷和鉀吸收利用相關性狀的QTL定位分析

2.4.1 葉片硝態氮 在白邑、玉溪各檢測到1個QTL,分布在第1、6染色體上,LOD值為3.55和4.11,加性效應為-53.05和97.02,增效基因分別來自十和田和麗江新團黑谷,貢獻率為35.19%、 27.07%。




2.4.5 氮素收獲指數 在玉溪、尋甸各檢測到1個QTL,分別位于第1、6染色體的RM3652-RM6324和RM541-RM7555區間,LOD值分別為3.51、3.09,加性效應為-3.02、-8.54,貢獻率分別為14.47%和31.85%,增效基因均來自十和田。
2.4.6 稻谷磷含量 僅在白邑點檢測到1個QTL,位于第1染色體RM3252-RM7180區間,LOD值為4.15,貢獻率為27.89%,加性效應為0.03,增效基因來自麗江新團黑谷。


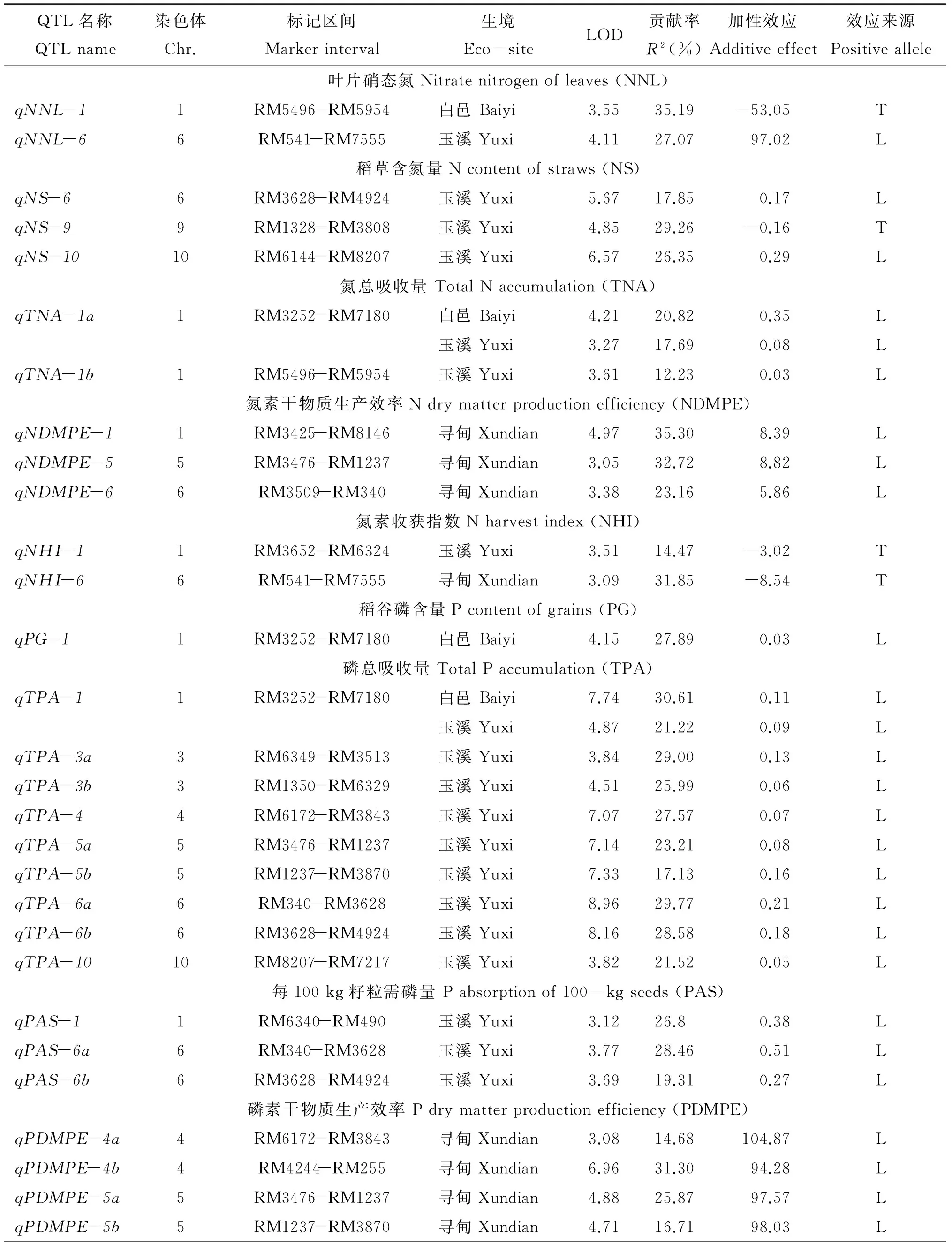
表5 低溫脅迫和正常生長環境下檢測到的氮、磷和鉀吸收利用相關性狀的主效QTLTable 5 Summary of QTLs for traits associated to N, P and K uptake utilization of NILs population detected under cold stress and normal conditions
續表5 Table 5 Continued

QTL名稱QTLname染色體Chr.標記區間Markerinterval生境Eco-siteLOD貢獻率R2(%)加性效應Additiveeffect效應來源PositivealleleqPDMPE-6a6RM340-RM3628尋甸Xundian4.3333.16109.57LqPDMPE-6b6RM3628-RM4924尋甸Xundian4.4229.70105.85L 磷素收獲指數Pharvestindex(PHI)qPHI-11RM3740-RM5302尋甸Xundian3.9910.633.65T玉溪Yuxi3.0412.54-4.74TqPHI-4a4RM4244-RM255尋甸Xundian9.4917.18-5.35TqPHI-4b4RM5879-RM317尋甸Xundian3.4110.60-3.68TqPHI-55RM3796-RM3476尋甸Xundian7.2914.83-4.77TqPHI-66RM340-RM3628尋甸Xundian11.2921.11-5.86T玉溪Yuxi14.0829.92-23.07TqPHI-7a7RM5711-RM6432尋甸Xundian5.3312.21-4.77TqPHI-7b7RM3753-RM7161尋甸Xundian3.6225.82-2.71T玉溪Yuxi3.7631.57-12.23TqPHI-1010RM1375-RM6673尋甸Xundian3.508.883.35L 稻谷鉀含量Kcontentofgrains(KG)qKG-11RM3252-RM7180白邑Baiyi5.6232.290.03L 稻草鉀含量Kcontentofstraws(KS)qKS-22RM5654-RM6911白邑Baiyi6.1130.81-0.16TqKS-77RM1048-RM6728白邑Baiyi4.5627.68-0.15T 鉀總吸收量TotalKaccumulation(TKA)qTKA-11RM297-RM3252白邑Baiyi3.5813.870.25L 每100kg籽粒需鉀量Kabsorptionof100-kgseeds(KAS)qKAS-33RM1350-RM6329尋甸Xundian3.5014.140.71LqKAS-55RM3796-RM3476尋甸Xundian7.0528.650.44LqKAS-6a6RM541-RM7555尋甸Xundian4.0325.180.48LqKAS-6b6RM340-RM3628尋甸Xundian5.7621.100.39LqKAS-99RM257-RM7390尋甸Xundian4.9134.57-0.50T 鉀素稻谷生產效率Kgrainproductionefficiency(KGPE)qKGPE-44RM4244-RM255尋甸Xundian4.0915.23-13.03TqKGPE-1010RM6673-RM6144尋甸Xundian3.4917.0712.11L 鉀素收獲指數Kharvestindex(KHI)qKHI-4a4RM255-RM3866尋甸Xundian4.159.25-2.44TqKHI-4b4RM6172-RM3843玉溪Yuxi3.0224.555.13LqKHI-55RM3476-RM1237尋甸Xundian6.7715.60-3.12TqKHI-66RM3628-RM4924尋甸Xundian8.3619.66-3.54L玉溪Yuxi3.1928.304.56LqKHI-77RM5711-RM6432尋甸Xundian4.0611.77-2.89TqKHI-1010RM1375-RM6673尋甸Xundian5.4013.872.43L
注(Note): L—增效基因來自麗江新團黑谷Efficiency gene expressed from Lijiangxintuanheigu; T—增效基因來自十和田Efficiency gene expressed from Towada.


2.4.11 稻谷鉀含量 僅在白邑檢測到1個QTL,位于第1染色體RM3252-RM7180區間的qKG-1,LOD值為5.62,貢獻率為32.29%,加性效應為0.03,增效基因來自麗江新團黑谷。
2.4.12 稻草鉀含量 僅白邑檢測到2個QTL,位于第2、7染色體RM5654-RM6911和RM1048-RM6728區間,LOD值為6.11、4.56,貢獻率分別為30.81%、27.68%,加性效應-0.16、-0.15。增效基因來自十和田。
2.4.13 鉀素總吸收量 僅在白邑檢測到1個QTL,qTKA-1位于第1染色體RM297-RM3252區間,LOD值為3.58,貢獻率為13.87%,加性效應為0.25,增效基因來自麗江新團黑谷。

2.4.15 鉀素稻谷生產效率 僅在尋甸檢測到2個QTL,位于第4、10染色體,LOD值分別為4.09和3.49,貢獻率為15.23%和17.07%,加性效應為-13.03、3.11。增效基因分別來自十和田和麗江新團黑谷。

3 討論與結論
3.1 QTL定位

3.2 控制各性狀的QTL比較
QTL檢測顯示,本研究有13個區間同時控制多個性狀,第1和第4染色體上有2個區間,第5和第6染色體上有3個區間,第3、7和10染色體上各1個區間。其中qNNL-1和qTNA-1b位于第1染色體RM5496-RM5954區間;qTNA-1a、qPG-1、qTPA-1和qKG-1位于第1染色體RM3252-RM7180區間;qTPA-3b和qKAS-3位于第3染色體RM1350-RM6329區間;qTPA-4、qPDMPE-4a和qKHI-4b位于第4染色體RM6172-RM3843區間;qPDMPE-4b、qPHI-4a和qKGPE-4位于第4染色體RM4244-RM255區間;qNDMPE-5、qTPA-5a、qPDMPE-5a和qKHI-5位于第5染色體RM3476-RM1237區間;qTPA-5b和qPDMPE-5b位于第5染色體RM1237-RM3870區間;qPHI-5和qKAS-5位于第5染色體RM3796-RM3476區間;qNNL-6、qNHI-6和qKAS-6a位于第5染色體RM541-RM7555區間;qNS-6、qTPA-6b、qPAS-6b、qPDMPE-6b和qKHI-6位于第6染色體RM3628-RM4924區間;qTPA-6a、qPAS-6a、qPDMPE-6a、qPHI-6和qKAS-6b位于第6染色體RM340-RM3628區間;qPHI-7a和qKHI-7位于第7染色體RM5711-RM6432區間;qPHI-10和qKHI-10位于第10染色體RM1375-RM6673區間。上述QTL富集區域,可能為部分區域重疊,或者為“一因多效”或“緊密連鎖”,初定位很難確定。但這種QTL成族現象,說明控制同一性狀的QTL可能不是隨機分布,而是在染色體上存在著控制某一特定性狀的QTL集中區域,同時證實N、P、K養分吸收存在復雜的交互作用。另外,本研究檢測到第1、第5和第6染色體的RM3252-RM7180、RM3796-RM3476、RM3628-RM4924標記區域的QTL與利用同一群體檢測孕穗期耐冷性QTL相同(未發表),且有4個位點位于前人定位的耐冷性QTL集中區域,即第3染色體RM1350-RM6329區間的qTPA-3b[36]、第4染色體RM255-RM3866區間的qKHI-4a[37]、第7染色體RM1048-RM6728區間的qKS-7[38]、第10染色體RM8207-RM7217區間的qTPA-10[39],表明控制水稻耐冷性與養分利用的QTL/基因間關系緊密。Zhang等[40]研究認為,麗江新團黑谷攜有冷調節基因COR,本研究利用相同供體親本的近等基因系,基因COR是否也具有養分吸收調節功能有待進一步研究。針對這13個重要的QTL富集區域將來可進行不同年份間重現性驗證,并通過構建相應的分離群體,借助于生物信息學手段,進一步開展精細定位研究,以分解這些QTL族或探索它們間的多效性關系,最終實現控制耐冷、養分吸收的QTL聚合育種,提高水稻耐冷、肥高效利用新品種的改良和選育效率。
[1] Chakravarthi B K, Naravaneni R. SSR marker based DNA fingerprinting and diversity studying rice (OryzasativaL)[J]. African Journal of Biotechnology, 2006, 5(9): 684-688.
[2] Mori M, Onishi K, Tokizono Yetal. Detection of a novel quantitative trait locus for cold tolerance at the booting stage derived from a troppicaljaponicarice variety silewah[J]. Breeding Science, 2011, 61(1): 61-68.
[3] Theocharis A, Clement C, Barka E A. Physiological and molecular changes in plants grown at low temperatures[J]. Planta, 2012, 235(6): 1091-1105.
[4] Zia M S, Salim M, Aslam M, Gill and Rahmatullah M A.Effect of low temperature of irrigation water on rice growth and nutrient uptake[J]. Journal of Agronomy and Crop Science, 1992, 173 (1): 22-31.
[5] 林海建, 張志明, 張永中, 等.作物氮、磷、鉀利用相關性狀的QTL定位研究進展[J]. 植物營養與肥料學報, 2010, 16 (3): 732 -743. Lin H J, Zhang Z M, Zhang Y Zetal. Advancement of QTL analysis for traits associated to N, P and K utilization[J]. Plant Nutrition and Fertilizer Science, 2010, 16 (3): 732-743.
[6] Cho Y I, Jiang W Z, Chin J H. Identification of QTLs associated with physiological nitrogen use efficiency in rice[J]. Molecular Cells, 2007, 23(1): 72-79.
[7] 方萍, 陶勤南, 吳平. 水稻吸氮能力與氮素利用率的QTLs及其基因效應分析[J]. 植物營養與肥料學報, 2001, 7(2): 159-165. Fang P, Tao Q N, Wu P. QTLs underlying rice root to uptake NH4-N and NO3-N and rice N use efficiency at seedling stage[J]. Plant Nutrition and Fertilizer Science, 2001, 7(2): 159-165.
[8] Wei D, Cui K H, Ye G Yetal. QTL mapping for nitrogen-use efficiency and nitrogen-deficiency tolerance traits in rice[J]. Plant and Soil, 2012, 359(2): 281-295.
[9] Tong H H, Chen L, Li W Petal. Identification and characterization of quantitative trait loci for grain yield and its components under different nitrogen fertilization levels in rice (OryzasativaL.)[J]. Molecular Breeding, 2011, 28(4): 495-509.
[10] Feng Y, Cao L Y, Wu W Metal. Mapping QTLs for nitrogen-deficiency tolerance at seedling stage in rice (OryzasativaL.)[J]. Plant Breeding, 2010, 129(6): 652-656.
[11] Obara M, Kajiura M, Fukuka Yetal. Mapping of QTLs associated with cytosolic glutamine synthetase and NADH-glutamate synthase in rice (OryzasativaL.)[J]. Journal of Experimental Botany, 2001, 52(359): 1209-1217.
[12] Bi Y M, Kant S, Clarke Jetal. Increased nitrogen-use efficiency in transgenic rice plants over-expressing a nitrogen-responsive early nodulin gene identified from rice expression profiling[J]. Plant Cell and Environment, 2009, 32(12): 1749-1760.
[13] Kurai T, Wakayama M, Abiko Tetal. Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions[J]. Plant Biotechnology Journal, 2011, 9(8): 826-837.
[14] Xu Y F, Wang R F, Tong Y Petal. Mapping QTLs for yield and nitrogen related traits in wheat: influence of nitrogen and phosphorus fertilization on QTL expression[J]. Theoretical and Applied Genetics, 2014, 127(1): 59-72.
[15] 穆平, 黃超, 李君霞, 等. 低磷脅迫下水稻產量性狀變化及其QTL定位[J]. 作物學報, 2008, 34(7): 1137-1142. Mu P, Huang C, Li J Xetal. Yield trait variation and QTL mapping in a DH population of rice under phosphorus deficiency[J]. Acta Agronomica Sinica, 2008, 34(7): 1137-1142.
[16] Ming F, Zheng X W, Mi G Hetal. Identification of quantitative trait loci affecting tolerance to low phosphorus in rice(OryzasativaL.)[J]. Chinese Science Bulletin, 2000, 45(6): 520-525.
[17] Hu B, Wu P, Liao C Yetal. QTLs and epistasis underlying activity of acid phosphatase under phosphorus sufficient and deficient condition in rice(OryzasativaL.)[J]. Plant and Soil, 2001, 230(1): 99-105.
[18] Yohei K, Juan P T, Terry Retal. QTLs for phosphorus deficiency tolerance detected in upland NERICA varieties[J]. Plant Breeding, 2013, 132(3): 259-265.
[19] Ni J J, Wu P, Senadhira D, Huang N. Mapping QTLs for phosphorus deficiency tolerance in rice (OryzasativaL.)[J]. Theoretical Applied Genetics, 1998, 97(8): 1361-1369.
[20] Koyama M L, Levesley A, Koebner R Metal. Quantitative trait loci for component physiological traits determining salt tolerance in rice[J]. Plant Physiology, 2001, 125(1): 406-422.
[21] Lin H X, Zhu M Z, Yano Metal. QTLs for Na+and K+uptake of the shoots and roots controlling rice salt tolerance[J]. Theoretical and Applied Genetics, 2004, 108(2): 253-260.
[22] Shimizu A, Guerta C Q, Gregorio G Betal. QTLs for nutritional contents of rice seedlings(OryzasativaL.) in solution cultures and its implication to tolerance to iron-toxicity[J]. Plant and Soil, 2005, 275(1): 57-66.
[23] 楊樹明, 王荔, 曾亞文, 等. 粳稻麗江新團黑谷近等基因系孕穗期耐冷性指標性狀的遺傳分析[J]. 華北農學報, 2013, 28(1): 7-11. Yang S M, Wang L, Zeng Y Wetal.Genetic analysis on cold tolerance characteristics at booting stage in the near-isogenic lines from Japonica rice Lijiangxintuanheigu[J]. Acta Agriculturae Boreali Sinica, 2013, 28 (1): 7-11.
[24] 曾亞文, 葉昌榮,申時全.水稻穗期耐冷NILs研制和QTL分析[J]. 中國農業科學, 2000, 33(4): 109. Zeng Y W, Ye C R, Shen S Q. Chromosomal Location of gene and development of near-isogenic lines for cold tolerance in rice[J].Scientia Agricultura Sinica, 2000, 33(4): 109.
[25] 李合生. 植物生理生化實驗原理與技術[M]. 北京: 高等教育出版社, 2000. 184-185. Li H S. Principles and techniques of plant physiological biochemical experiment[M]. Beijing: Higher Education Press, 2000. 184- 185.
[26] 鮑士旦. 土壤農化分析[M]. 北京: 北京農業大學出版社, 2000. 30-107. Bao S D. Soil agrochemistry analysis[M]. Beijing: Beijing Agricultural University Press, 2000. 30-107.
[27] Jiang L G, Dai T B, Jiang Detal. Characterizing physiological N-use efficiency as influenced by nitrogen management in three rice cultivars[J]. Field Crops Research, 2004, 88(2): 239-250.
[28] Murray M G,Thompson W F. Rapid isolation of high molecular weight plant DNA[J]. Nucleic Acids Research, 1980, 8(19): 4321-4325.
[29] Temnykh S, Park W D, Ayres Netal. Mapping and genome organization of microsatellite sequences in rice (OryzasativaL.)[J]. Theoretical and Applied Genetics, 2000, 100(5): 697-712.
[30] 王建康. 數量性狀基因的完備區間作圖方法[J]. 作物學報, 2009, 35 (2): 239-245. WANG J K. Inclusive composite interval mapping of quantitative trait genes[J]. Acta Agronomica Sinica, 2009, 35 (2): 239-245.
[31] Mccouch S R. Gene nomenclature system for rice[J]. Rice, 2008, 1(1): 72-84.
[32] Dong W, Ke H C, Jun F Petal. Genetic dissection of grain nitrogen use efficiency and grain yield and their relationship in rice[J]. Field Crops Research, 2011, 124(3): 340-346.
[33] Wan X Y, Wan J M, Weng J Fetal. Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments[J]. Theoretical and Applied Genetics, 2005, 110(7): 1334-1346.
[34] Wissuwa M, Yano M, Ae N. Mapping of QTLs for phosphorus deficiency tolerance in rice (OryzasativaL.)[J]. Theoretical and Applied Genetics, 1998, 97(6): 777-783.
[35] Wang C, Ying S, Huang Hetal. Involvement ofOsSPX1 in phosphate homeostasis in rice[J]. Plant Journal, 2009, 57(5): 895-904.
[36] Shirasawa S, Endo T, Nakagomi Ketal. Delimitation of a QTL region controlling cold tolerance at booting stage of a cultivar, ‘Lijiangxintuanheigu’, in rice,OryzasativaL[J]. Theoretical and Applied Genetics, 2012, 124(5): 937-946.
[37] Saito K, Hayano S Y, Kuroki M, Sato Y. Map-based cloning of the rice cold tolerance geneCtb1 [J]. Plant Science, 2010, 179(1): 97-102.
[38] Zhou L, Zeng Y W, Hu G Letal. Characterization and identification of cold tolerant near-isogenic lines in rice[J]. Breeding Science, 2012, 62(2): 196-201.
[39] Ye C, Fukai S, Godwin I Detal. A QTL controlling low temperature induced spikelet sterility at booting stage in rice[J]. Euphytica, 2010, 176(3): 291-301.
[40] Zhang T, Zhao X, Wang W Setal. Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes[J]. PloS One, 2012, 7(8): e43274.
Identification of QTL traits on N, P and K utilization in rice under different growth environments
YANG Shu-ming1,2, ZENG Ya-wen1,3*, WANG Li4*, DU Juan1, PU Xiao-ying1, YANG Tao1
(1BiotechnologyandGeneticResourcesInstitute,YunnanAcademyofAgriculturalSciences,Kunming650205,China;2KeyLaboratoryoftheSouthwesternCropGeneResourcesandGermplasmInnovation,MinistryofAgriculture,Kunming650223,China; 3AgriculturalBiotechnologyKeyLaboratoryofYunnanProvince,Kunming650223,China;4CollegeofAgronomyandBiotechnology,YunnanAgriculturalUniversity,Kunming650201,China)
【Objectives】To provide the basis for the breeding program of molecular assistant selection (MAS) and map-based cloning of high nutrition efficiency utilization, the traits of quantitative trait locus (QTLs) on nitrogen, phosphorus and potassium utilization in rice were identified. 【Methods】 A set of 105 near-isogenic lines, BC4F8population was developed by backcrossing between ‘Lijiangxintuanheigu’ (the stongly cold-tolerantjaponicalandrace, grant No.2) as a donor parent and ‘Towada’ (cold-sensitivejaponicacultivar) as a recurrent parent, and was used as materials, and were planted in Baiyi (cold water irrigation), Xundian (natural low temperature condition) and Yuxi (normal growing environment) in Yunnan, respectively. Sixteen traits associated with N, P, K utilization were evaluated under three different ecological environments. The phenotypic data of 105 NILs, genetic map containing 180 microsatellite markers with 1820.6 cM of the genome, and 15.67 cM average distance between the markers were used to analyze QTLs of N, P and K utilization in rice by the statistic software of QTL IciMapping V 3.2. 【Results】 The total 56 QTLs were detected under three different environment, and were confirmed to be distributed on chromosome 1, 2, 3, 4, 5, 6, 7, 9, and 10, respectively. The number of QTL for single trait was varied from 1 to 10, and the single QTL accounted for 8.88%-35.30% of the phenotypic variation. N, P and K utilization efficiency of No.12, 27, and 17 QTLs were respectively detected. Six QTLs,qTNA-1a,qTPA-1,qPHI-1,qPHI-6,qPHI-7b, andqKHI-6 were detected under cold stress and normal condition, and had high stability and explained 10.63%-31.57% of the phenotypic variation. Moreover, QTLs showed the cluster distribution in 13 QTL regions of chromosome 1, 3, 4, 5, 6, 7, and 10, and a single QTL controlled 2-5 traits, and most of these traits co-controlled total P accumulation, P dry matter production, P harvest index, K absorption of 100-kg seeds, and K harvest index. 【Conclusions】 In this study, 56 QTLs related to N, P and K utilization in rice were detected, and the QTLs with high contribution might be useful for rice breeding with high nutrition efficiency utilization for N, P and K by MAS. Additionally, 13 genomic regions of QTLs cluster distribution are important candidate regions for further study.
rice (OryzasativaL.); near-isogenic lines; nutrient uptake; quantitative trait locus; QTL pleiotropy
2014-07-03 接受日期: 2014-10-21 網絡出版日期: 2015-05-20
云南省技術創新人才培養項目(2011CI059和2012HB050);云南省科技惠民計劃項目(2014RA060)資助。
楊樹明(1973—),男,云南武定人,博士,研究員,主要從事農作物種質資源高效基因挖掘、遺傳育種研究。 Tel: 0871-65894145,E-mail: yangshuming126@126.com。* 通信作者 E-mail: zengyw1967@126.com; E-mail: 13759505639@163.com
S511.01
A
1008-505X(2015)04-0823-013

