Evaluation of polygenic risk score for risk prediction of gastric cancer
Xiao-Yu Wang, Li-Li Wang, Lin Xu, Shu-Zhen Liang, Meng-Chao Yu, Qiu-Yue Zhang, Quan-Jiang Dong
Xiao-Yu Wang, Li-Li Wang, Lin Xu, Shu-Zhen Liang, Meng-Chao Yu, Quan-Jiang Dong, Central Laboratories and Department of Gastroenterology, Qingdao Municipal Hospital, Qingdao University, Qingdao 266071, Shandong Province, China
Qiu-Yue Zhang, Department of Clinical Laboratory, the Eighth Medical Center of the General Hospital of the People’s Liberation Army, Beijing 100000, China
Abstract Genetic variations are associated with individual susceptibility to gastric cancer. Recently, polygenic risk score (PRS) models have been established based on genetic variants to predict the risk of gastric cancer. To assess the accuracy of current PRS models in the risk prediction, a systematic review was conducted. A total of eight eligible studies consisted of 544842 participants were included for evaluation of the performance of PRS models. The overall accuracy was moderate with Area under the curve values ranging from 0.5600 to 0.7823. Incorporation of epidemiological factors or Helicobacter pylori (H. pylori) status increased the accuracy for risk prediction, while selection of single nucleotide polymorphism (SNP) and number of SNPs appeared to have little impact on the model performance. To further improve the accuracy of PRS models for risk prediction of gastric cancer, we summarized the association between gastric cancer risk and H. pylori genomic variations, cancer associated bacteria members in the gastric microbiome, discussed the potentials for performance improvement of PRS models with these microbial factors. Future studies on comprehensive PRS models established with human SNPs, epidemiological factors and microbial factors are indicated.
Key Words: Polygenic risk scores; Gastric cancer; Helicobacter pylori; Gastric microbiome
INTRODUCTION
Gastric cancer (GC) is the fourth most commonly diagnosed type of cancer worldwide and the second leading cause of cancer-related death[1]. According to the latest cancer statistics, there were approximately 26380 new gastric cancer cases and 11090 deaths in the United States in 2022[2]. The occurrence of gastric cancer results from a combination of risk factors, including host genetic factors andHelicobacter pylori(H. pylori) infection[3,4]. Genetic variations play an important role in the occurrence and progression of gastric cancer[5,6]. Genome-wide association studies have identified many single nucleotide polymorphisms (SNPs) in the human genome that are involved in the development of gastric cancer.H. pyloriinfection affects approximately half of the world’s population. The pathogen is considered a definite carcinogen of gastric cancer[7]. Epidemiological studies have revealed that age, sex, alcohol consumption and smoking are risk factors for gastric cancer[8].
To prevent the development of gastric cancer, it is important to identify individuals at high risk for cancer and apply intervention measures to impede the progression of the disease. Many studies have been conducted to explore the performance of biomarkers or models established with risk factors for predicting gastric cancer risk. Models based on epidemiological factors, including age, sex andH. pyloriinfection, have adequate performance in the prediction of gastric cancer risk[9,10]. Genetic variations ofH. pylorishow great potential for use in the prediction of gastric cancer risk[11]. Cancer-associated SNPs have been reported to be valuable in stratifying gastric cancer risk based on the genetic background[12,13].
Polygenic risk score (PRS) models are established with a number of SNPs or genetic variants to explore the combined effect of multiple genetic variations in the risk prediction of disease[14]. They show improved performance in the prediction of the risk of breast, prostate, and colorectal cancer and diseases involving multiple genetic factors[15-17]. Calculations of PRS vary among different models. The simplest way to calculate the PRS is summing the number of all risk alleles[18]. Considering variations in the cancer risk associated with different SNPs, each risk allele is weighted by its odds ratio (OR) value for cancer. PRS is then calculated as a sum of weighted risk alleles[14]. To date, studies have been conducted employing PRS models to predict gastric cancer risk. Different sets of cancer-associated SNPs have been used in the PRS models. In certain studies, epidemiological factors have been included in the establishment of the models. To assess the performance of PRS models in the prediction of gastric cancer risk, this article aimed to comprehensively analyze the accuracy of PRS models for risk prediction through a systematic review of related studies and discuss potentials in the performance improvement of PRS models with the inclusion ofH. pylorigenetic variations and bacterial members of the gastric microbiome for use in the future.
PERFORMANCE OF CURRENT PRS MODELS
To assess the performance of PRS models for the prediction of gastric cancer risk, a systematic review was conducted. The details of the methods for the systematic review can be found in Supplementary material. The PRISMA flow chart (Figure 1) shows the article retrieval and filtering process. A total of 165 articles were retrieved from PubMed, Web of Science, and Embase electronic database using the search strategies in the Methods (Supplementary material). After removing duplicates, a total of 96 articles were further screened. According to the topics and abstracts, 37 articles were selected and analysed for eligibility. Of them, 8 studies were eligible and included in our systematic review in accordance with the inclusion and exclusion criteria[19-26]. The reasons for exclusion are shown in the PRISMA flow diagram.
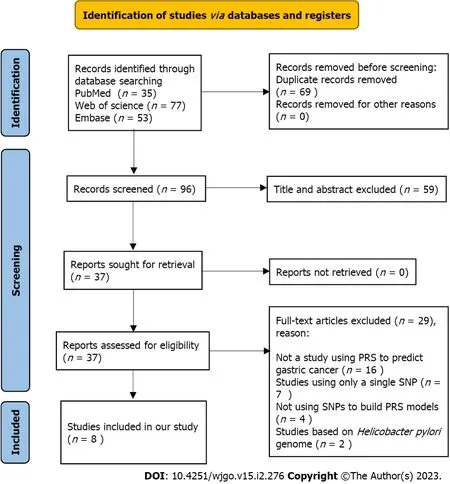
Figure 1 PRISMA flow diagram. PRISMA flow diagram showed the process of the study selection. SNP: Single nucleotide polymorphism; PRS: Polygenic risk score.
The characteristics of the included studies are shown in Table 1. All of the studies were published in the last five years. Six of them were case-control studies, and two were cohort studies. The study areas included China (5), Korea (1), Japan (1) and Europe (1). The sample sizes in the included studies ranged from 1088 to 400807. A total of 544842 participants were included. All the studies established a PRS model to predict the risk of gastric cancer.
Details regarding the establishment and evaluation of PRS models in these studies are described in Table 2. The accuracy of the PRS models was assessed with the Area under the curve (AUC) in five included studies. The performance of the models was moderate, with AUC values ranging from 0.5600 to 0.7823. The highest performance has been reported by Ishikuraet al[25] from Japan, with an AUC of 0.7677 in the training set and 0.7823 in the validation set. Two studies from China have used ORs to evaluate the performance of PRS models[19,23]. OR values for the highest quartile with respect to the lowest are 1.14 and 1.19, indicating that the performance of the models was unsatisfactory. The hazard ratio (HR) was used in the remaining study with a value of 2.08 for the highest quantile with respect to the lowest. Overall, the performance of the current PRS models varies considerably among the included studies and appears to have a moderate predictive power for gastric cancer. Factors affecting the performance are indicated.
Pearson correlation analysis of sample sizes and AUC values demonstrated that there was no significant correlation between them (r = -0.51,P= 0.380). This result suggested that the variations in the sample size of the included studies had minimal influence on the predictive power of the PRS model. The number of genetic variants in the models ranged from 3 to 112 SNPs. Pearson correlation analyses were performed to explore whether the number of SNPs was related to the predictive power of the model. The results showed that there was no significant correlation between the number of SNPs and AUC (r= 0.85,P= 0.067). Our results are consistent with previous systematic reviews of breast cancer[27]. This suggests that the inclusion of more SNPs in the PRS model would not improve the performance in the prediction of gastric cancer risk.
All of the included studies used the weighted PRS method instead of the simple counting method. The weight of risk alleles is crucial for the performance of the PRS models[14,28]. Of note, the same SNP, such as rs2294008, has been used in different models, but the weights varied greatly (Table 1). Accordingly, the predictive power varied among studies (Table 2). Generally, the weight of a risk allele derives from the OR of risk alleles for the development of gastric cancer. In the included studies, the ORs mostly came directly from the results of case-control comparisons. They have not been, however, confirmed in a validation set. This might account for the different weights that have been used among studies. It appears that a validation of the OR is required to eliminate the bias of weights among studies, improving the consistency in the performance of PRS models.
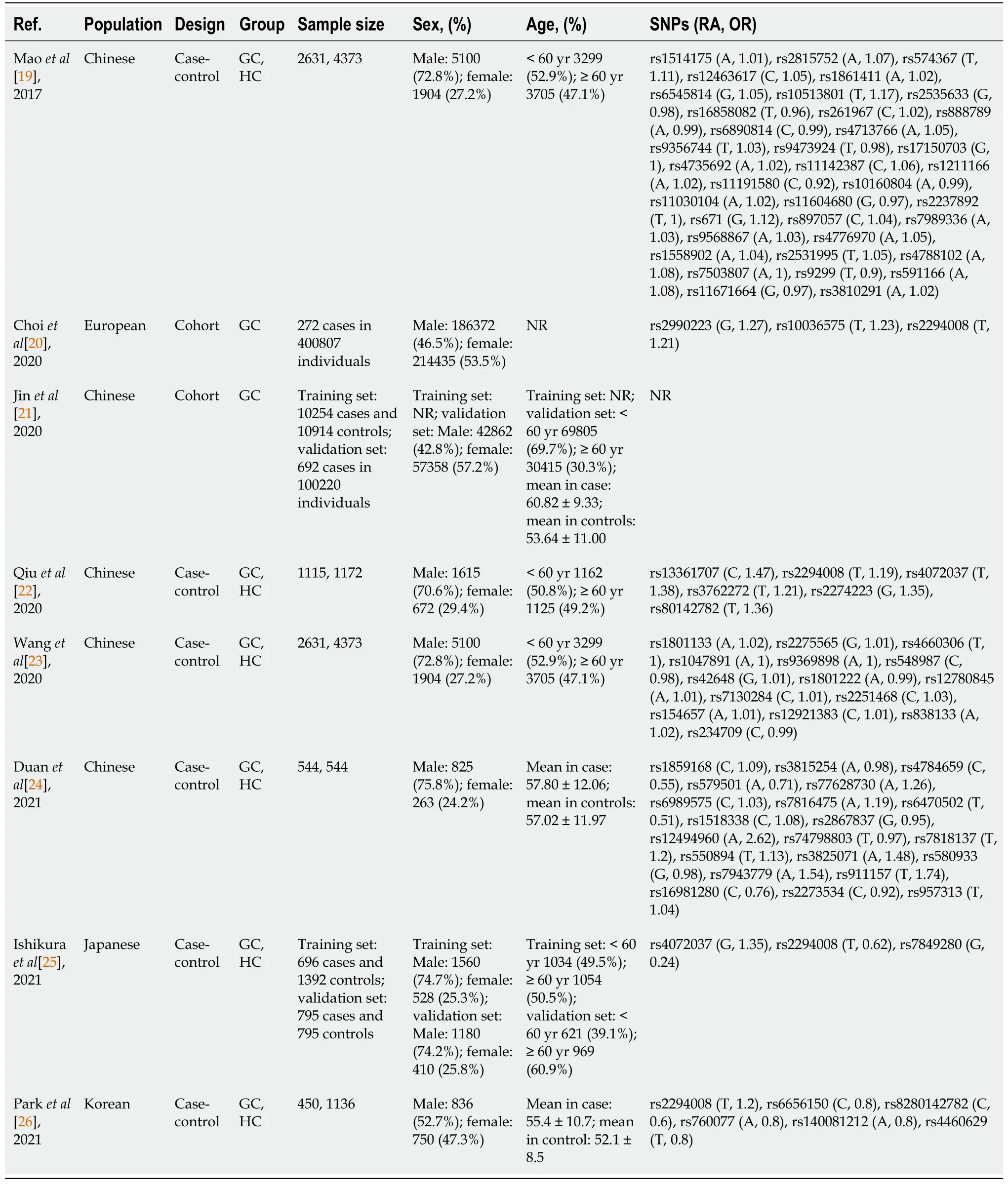
Table 1 Main characteristics of the included studies
To improve the accuracy of the PRS model in the prediction of cancer risk, certain epidemiological factors implicated in cancer development have been considered[29,30]. A number of epidemiological factors are associated with the occurrence of gastric cancer. Individuals of male sex and older age are at increased risk for gastric cancer[9,31]. Previous studies have revealed environmental factors in gastric cancer development. A family history has been considered to be significantly associated with the occurrence of gastric cancer, with OR of more than 2[32]. Studies have revealed a close association between alcohol consumption and the risk of gastric cancer. A meta-analysis showed that heavy alcohol consumption increases the risk of gastric cancer[33,34]. Smokers have been reported to be at high risk of gastric cancer[35]. In this study, epidemiological factors were included in five studies in addition to genetic variations (Table 2). After taking into account the epidemiological factors, the AUC achieved using each model increased by 0.014 to 0.215. To explore whether the predictive performance of the PRS model was improved after epidemiological factors were included, the Mann-Whitney test was performed. The results showed that the AUC values were significantly increased from 0.56-0.74 to 0.61-0.78 after epidemiological factors were considered (P= 0.047).
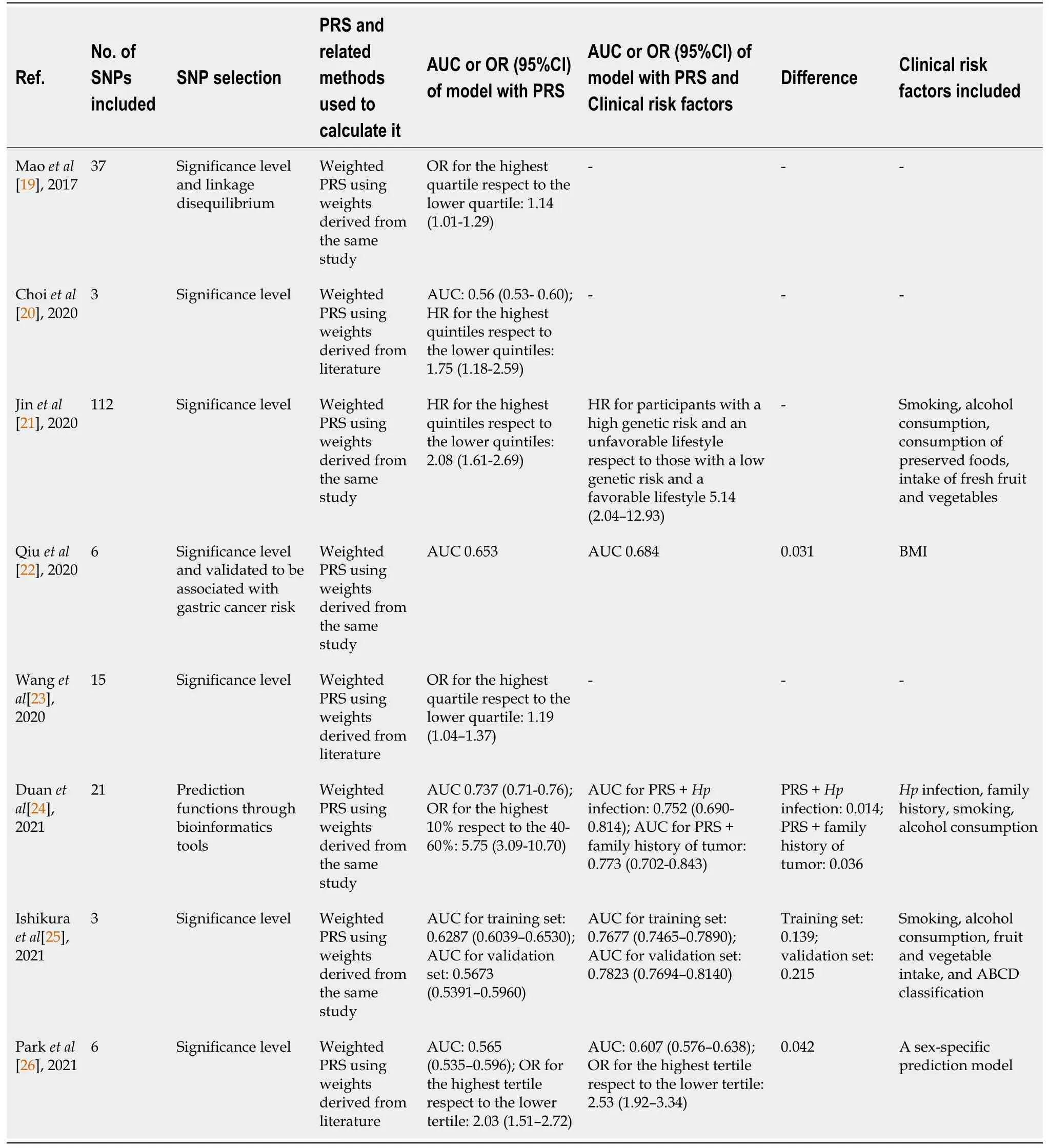
Table 2 Development and evaluation of Polygenetic risk scores for predicting gastric cancer
H. pyloriis a major cause of gastric cancer. The risk of non-cardia gastric cancer inH. pylori-infected individuals is 6 times higher than that in uninfected individuals[36]. Only one of the included studies tookH. pyloriinto account in the establishment of the PRS model. The performance of the model for the prediction of gastric cancer risk increased with an AUC value increasing from 0.737 to 0.752.H. pyloriinfection serves as a biomarker for gastric cancer and has been combined with other epidemiological factors to predict gastric cancer risk. Tanet al[37] found thatH. pyloriinfection alone had moderate power for predicting gastric cancer risk with an AUC of 0.66. The accuracy of prediction was improved after other clinical factors were incorporated. A study consisting of 14929 participants demonstrated thatH. pyloriinfection combined with seven epidemiological factors has a high predictive power with an AUC value of 0.76[38]. These findings suggest that incorporatingH. pyloristatus into PRS models boosts the predictive power for gastric cancer risk.
In many types of cancer, PRS models have shown great power for risk prediction[39,40]. Our analyses demonstrated that the performance of current PRS models is promising in predicting the risk of gastric cancer. Nonetheless, the predictive power is not as satisfying as expected. Inclusion of epidemiological factors andH. pyloriinfections likely enhances the performance of the PRS model for the prediction of gastric cancer risk.
RISK PREDICATION WITH MICROBIAL FACTORS
In the reported PRS models, the risk of gastric cancer has been predicted mainly based on the genetic susceptibility resulting from genetic variations and common cancer risk factors, including age, sex, smoking status, and alcohol consumption. Previous studies have reported serum pepsinogen status could reflect the extent of atrophic change in gastric mucosa[41]. The combination of serum pepsinogen status andH. pyloristatus serves as a valuable marker for stratifying the risk of gastric cancer[42]. Individuals with decrease status of pepsinogen andH. pyloriinfection had a higher risk of gastric cancer compared with healthy control, with a HR value of 6.0[43]. Furthermore, recent studies have demonstrated that many microbial factors play roles in gastric carcinogenesis. Infection withH. pyloricauses gastric cancer in only a minority of individuals[44]. Genetic differences between strains ofH.pyloriaccount in part for the differential outcomes of the infection among individuals[45]. The dysbiotic gastric microbiome plays an important role in the development of gastric cancer[46,47]. In addition, studies have shown that other bacteria may play an important role in promoting cancer, following the structural imbalance of the stomach microbiome induced byH. pylori[48,49].As we mentioned, multiple studies have reported other bacteria that are associated with gastric cancer[50,51]. We believe that the gastric microbiome can be used as a valuable candidate to establish a prediction model for the occurrence of gastric cancer.
Gastric cancer-associated SNPs of H. pylori
The genome ofH. pyloriis substantially diverse[45]. There is a high level of differences in the gene contents, deletion/insertion, genetic inversion, sequence variations and SNPs[52]. Genetic variations in virulence genes, includingcagA,vacA, andbabA,are closely associated with gastric cancer risk[53,54]. Genome-wide association studies have identified a number of gastric cancer-associated SNPs in theH.pylorigenome[55,56]. These cancer-associated genetic variations ofH. pylorican be used in the risk prediction of gastric cancer. During the process of screening the studies (Figure 1), we observed that studies used gastric cancer-associated SNPs of theH. pylorigenome to predict gastric cancer risk. Using a model comprising six validated loci in thecagpathogenicity island, a study on 1220 subjects demonstrated a sound predictive power for gastric cancer with an AUC of 0.65[57]. Berthenetet al[55] generated a risk score model with 12 gastric cancer-associated SNPs identified by a GWAS study ofH.pylori.The results of this study have shown that the model is capable of predicting gastric cancer risk. A recent report established a PRS model with gastric cancer-associated SNPs selected from previous studies[11]. The model based onH. pyloriSNPs achieved good predictive performance. These results convincingly support that the incorporation ofH. pylorigenomic variations into current PRS models would considerably enhance the accuracy in the prediction of gastric cancer risk.
Cancer-associated bacteria in the gastric microbiome
Dysbiosis of the gastric microbiome promotes the development of gastric cancer[46,47]. Many bacteria in the gastric microbiome possess carcinogenic potential[50,51,58].
An observational study of 1043 patients demonstrated a significant enrichment ofStreptococcus anginosus(S. anginosus) andStreptococcus constellatus(S. constellatus) in gastric cancer[58,59]. The abundances ofS. anginosusandS. constellatusserve as novel faecal signatures of early gastric cancer. Cokeret aldemonstrated an association betweenS. anginosus,Peptostreptococcus stomatis,Parvimonas micra,Slackia exigua, Dialister pneumosintesand gastric cancer[60]. These bacteria could form a synergistic network, leading to additional contributions to the disease. They could be used as potential tissue markers for gastric cancer with AUC values of 0.82 and 0.81 in the discovery and validation cohorts, respectively. Pnget al[61] conducted a cohort study involving 43 participants to identify potential carcinogenic bacteria. The study demonstrates that theMoryellagenus,Vibrogenus,Comamonadaceaefamily,Paludibactergenus,Agrobacteriumgenus, andClostridialesorder in the gastric microbiome are associated with gastric cancer. The model containing analyses of these bacteria is capable of predicting early gastric cancer with an AUC of 0.82. It has been shown that a random forest model generated with bacterial members of the gastric microbiome has a high performance in risk prediction[62,63]. These findings collectively support that bacterial members of the gastric microbiome have potential in the risk stratification of gastric cancer. Despite the requirement of further validation, the inclusion of the analysis of these bacteria in PRS models most likely enhances the accuracy in the prediction of gastric cancer.
CONCLUSION
Our systematic review showed that PRS models have great potential in the prediction of gastric cancer. Incorporation of other risk factors for gastric cancer could increase the accuracy of the models. To further increase the predictive performance of PRS models for gastric cancer, a comprehensive PRS model generated with the analysis of epidemiological risk factors, genetic variations ofH. pylori,and bacterial members of the gastric microbiome in addition to human genetic variations requires further evaluation. PRS models with high accuracy would benefit the development of individual risk scores, facilitating the prevention of gastric cancer.
FOOTNOTES
Author contributions:Wang XY and Liang SZ collected sequencing data; Wang LL and Xu L analyzed the data; Wang XY, Yu MC, and Zhang QY wrote the manuscript; Dong QJ designed the research and supervised the manuscript; all authors reviewed the manuscript and approved the final version of the manuscript.
Supported bythe National Natural Science Foundation of China, No. 31870777.
Conflict-of-interest statement:The authors declare that they have no competing interests.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:China
ORCID number:Xiao-Yu Wang 0000-0002-3278-0879; Li-Li Wang 0000-0002-3607-0786; Lin Xu 0000-0003-3098-8251; Quan-Jiang Dong 0000-0002-5226-7853.
S-Editor:Chen YL
L-Editor:A
P-Editor:Chen YL
 World Journal of Gastrointestinal Oncology2023年2期
World Journal of Gastrointestinal Oncology2023年2期
- World Journal of Gastrointestinal Oncology的其它文章
- Comment on “Crosstalk between gut microbiota and COVID-19 impacts pancreatic cancer progression”
- Prognostic value of claudin 18.2 expression in gastric adenocarcinoma
- Potent bromodomain and extraterminal domain inhibitor JAB-8263 suppresses MYC expression and exerts anti-tumor activity in colorectal cancer models
- microRNA-627-5p inhibits colorectal cancer cell proliferation,migration and invasion by targeting Wnt2
- Increased CD4/CD8 Lymphocyte ratio predicts favourable neoadjuvant treatment response in gastric cancer: A prospective pilot study
- Cancerous inhibitor of protein phosphatase 2A enhances chemoresistance of gastric cancer cells to oxaliplatin
