MiR-36f對豬蛔蟲幼蟲感染力和發育的影響
馮勝勇,付京花,2,邵長春,3,朱興全,徐民俊,2
MiR-36f對豬蛔蟲幼蟲感染力和發育的影響
馮勝勇1,付京花1,2,邵長春1,3,朱興全1,徐民俊1,2
目的 探討豬蛔蟲蟲卵和幼蟲期特異表達的miR-36f的是否與豬蛔蟲幼蟲的感染性和發育有關。方法 將合成的miR-36f的模擬物,抑制劑以及模擬物對照和抑制劑對照通過浸泡的方法被豬蛔蟲三期幼蟲吸收后,分別將這四組幼蟲通過灌胃的方式感染小鼠,以不作任何處理的幼蟲感染小鼠作為空白對照組,四天后采用貝爾曼法收集各組小鼠肺和肝的幼蟲。對各組回收幼蟲的數量以及幼蟲的長度和寬度進行測量,并采用qRT-PCR方法對miR-36f的兩個靶基因OST48和cytochrome b進行定量分析。結果 抑制miR-36f以后,miR-36f的兩個靶基因OST48和cytochrome b均被下調,幼蟲發育和感染力受到抑制。結論 提示豬蛔蟲的miR-36f與幼蟲個體發育和感染力有關。
豬蛔蟲;asu-miR-36f;感染力;發育
Liu Wen-ge and Xu Xiao-pei contributed equally.
Toxoplasmagondii, the etiologic agent of toxoplasmosis,can spread to all warm-blooded animals, including birds and humans around the world[1-2]. Despite the majority cases ofToxoplasmainfection is asymptomatic in immunocompetent humans,serious manifestations including encephalitis, myocarditis or hepatitis may occur in immunocomprised individuals[3-4], and the parasite can also cause severe diseases in pregnant women[5-6]. Also,T.gondiiis responsible for major economic losses of livestock industry, due to the causes of abortion or congenital toxoplasmosis of domestic animals, especially of sheep and goats[7- 8].
Recent research suggests that different clonal types amongT.gondiistrains with diverse geographical locations could cause different consequences in livestock and humans[9-10]. Therefore, a deep insight of genetic diversity ofT.gondiiis central for better understanding epidemiological patterns and pathogenicity, as well as exploring the new strategies for vaccination, diagnosis, or treatment of toxoplasmosis.
Microneme proteins (MICs), hiding in the micronemes ofT.gondii, play an important role in host cell attachment and the parasite actomyosin[11]. MIC5 is a member of microneme proteins, directly regulating the activity of proteases, and also it is related to the proteolytic susceptibility of other microneme protein substrates[12].
Little,however,has been known of the variation of MIC5 gene sequence inT.gondiiisolates. The present study aims to reveal the sequence variation of MIC5 gene based on the examined sequences of MIC5 gene among 13T.gondiistrains from different hosts and geographical locations.
Materials and methods
T.gondiiisolates
A total of 13T.gondiistrains(belonging to different genotypes)from different hosts and geographical locations were used for analysis in this research (Table 1), and genomic DNAs (gDNA) were extracted as described previously[13-14].
According to the nucleotide sequence of MIC5 (GT1) gene provided by ToxoDB Database (TGGT1-277080), a pair of specific primers (forward primer:5′-ATGCTGCGACCTACTGTTCGTA-3′;and reverse primer: 5′-CTATGCGAGTTTCACCTCGGAG-3′) were used to amplify MIC5 gene from each isolate. The amplification reaction was performed in a total volume of 25 μL containing 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, 250 μM each of dNTP, 0.2 μM of each primer, 100-200 ng of template DNA, and 0.25 UExTaqpolymerase (TaKaRa). The PCR program was executed in a thermorcycle (Biometra) under the following conditions: after an initial denaturation at 95 ℃ for 5 min, followed by 30 cycles of 95 ℃ for 1 min (denaturation), 60.5 ℃ for 45 s (annealing), 72.0 ℃ for 2 min(extension), finally, an additional extension at 72 ℃ for 10 min. Each amplicon (5 μL) was confirmed by electrophoresis on a 1% (w/v) agarose gel, stained with GoldenViewTMand photographed using a gel documentation system (UVP GelDoc-ItTMImaging System, Cambridge, UK.)[15].

Tab.1 Details of Toxoplasma gondii strains used
Note: * based on the results of[6, 13-14]
The MIC5 PCR products were purified by spin columns (WizardTMPCR-Preps DNA Purification System, Promega, USA), and ligated with pMD 18-T vector according to manufacturer’s recommendations (TaKaRa). Then, the recombinant vectors were transformed into the JM109 competent cells (Promega, USA).After screening by PCR amplification and enzyme digestion, positive cloning was sequenced by Shanghai Songon Biotechnology Company[16].
The sequences of MIC5 gene from differentT.gondiistrains were aligned using Multiple Sequence Alignment Program, ClustalX 2.11[17],which was used to determine the sequence variation among the examinedT.gondiistrains.
Three referred methods, namely maximum parsimony (MP),Bayesian inference (BI) and maximum likelihood (ML), were used for phylogenetic re-construction.BI analysis was conducted with four independent Markov chains run for 10 000 000 metropolis coupled MCMC generations, sampling a tree every 10 000 generations in MrBayes 3.1.1[18]. The first 250 trees were omitted as burn-in and the remaining trees were used to calculate Bayesian posterior probabilities(PP).ML and MP analysis was performed using PAUP* 4.0b10[19], and Bootstrap support for ML trees was calculated using 100 bootstrap replicates and MP was calculated from 1 000 bootstrap replicates with 10 random additions per replicate[20].
Results and discussion
The MIC5 genesequences were 1 975 bp in length for 13 examinedT.gondiistrains, and their A+T contents were from 51.70% to 51.95%. Pairwise comparison based on the obtained sequences revealed nucleotide polymorphisms at 52 sequence variation positions (0-1.3%) (Table 2), including 38 variations in 3 introns, and 14 variations in 4 exons.These results indicated a low sequence variation in MIC5 gene. Moreover, a total of 40 transitions (C< >T/A< >G) and 12 transversions (A< >T/T< >G/C< >G/A< >C) in MIC5 genomic sequences were identified; Of which, 10 transitions (C< >T/A< >G) and 4 transversions(C< >T/A< >G)were in MIC5 exons regions, while 30 transitions (C< >T/A >G) and 8 transversions(A< >T/T< >G/C< >G/A< >C) in MIC5 intron regions. These results were similar to that of eIF4A, ROP9, MIC13 and PLP1[16, 21-23].
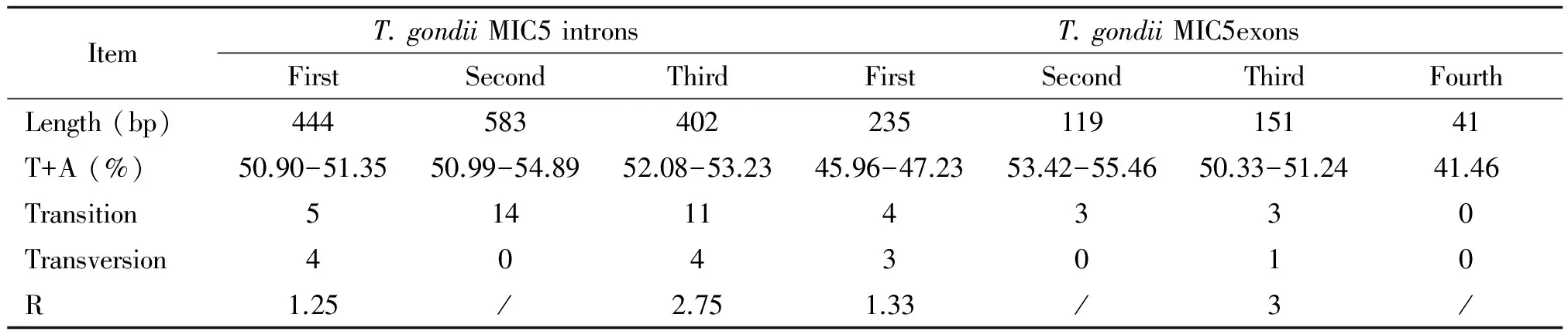
Tab.2 Characteristics of intron and exon sequences of Toxoplasma gondii MIC5gene
Note: R=transition/transversion.
Sequence variations in MIC5 between the three major genotypes were also calculated.When MIC5 sequences of Type I strains were compared with that of Type II strains, while there was no fixed nucleotide mutations in Type I strains, there were 13 fixed nucleotide variations in Type II strains; of which, 4 were transversions(C< >G/C< >A/G< >T) and 9 were transitions (C< >T/A< >G). When MIC5 sequences of Type I strains were compared with that of the Type III strain, there were 14 fixed nucleotide mutations in the Type III strain; Of which, 4 were transversions (C< >G/C< >A/G< >T) and 10 were transitions (C< >T/A< >G). When MIC5 sequences of Type II strains were compared with that of the Type III strain, there was 1 fixed mutation (transition A< >G) in the Type III strain.
The reconstructed phylogenetic tree (Figure 1) based on the MIC5 sequences using BI, MP and ML methods showed that one classical genotype (Type I) were able to be differentiated from the other two classical genotypes (Type II and III), but the type II and type III cannot entirely be separated from each other due to low sequence diversity,which was different from that using GRA5 gene sequences[20].
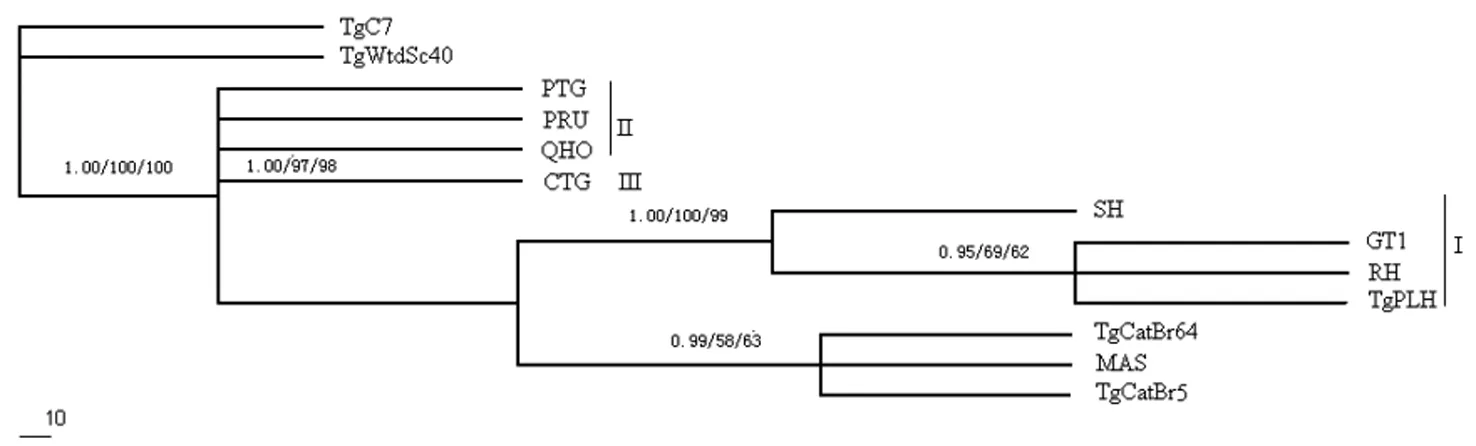
Classical genotype (Type I, II and III) were denoted by I, II, and III;
The phylogenetic tree was built by Bayesian inference (BI), maximum likelihood(ML)and maximum parsimony (MP) analysis;
The numbers on branches signify bootstrap values by BI/ML/MP.
Fig.1 Reconstruction of phylogenetic relationships of 13Toxoplasmagondiiisolates based on the whole sequences of the MIC5 gene
In conclusion, the present study revealed a high sequence homology in the entire MIC5 gene among 13T.gondiiisolates (belonging to different genotypes) from different regions and hosts, suggesting that MIC5 gene is not a marker appropriate for population genetic study ofT.gondiistrains.
[1]Dubey JP. Toxoplasmosis of animals and humans[M].2nd ed. New York:CRC Press Inc.,2010: 1 313.
[2]Dubey JP. The history ofToxoplasmagondii--The first 100 years[J]. J Eukaryot Microbiol,2008, 55: 467-475.DOI: 10.1111/j.1550-7408.2008.00345.x
[3]Bowie WR, King AS,Werker DH. Outbreak of toxoplasmosis associated with municipal drinking water[J]. Lancet, 1997, 350:173-177.
[4]Do HY. Clinical features[M].In: Human toxoplasmosis.Oxford:Oxford University Press, 1992: 56-78.
[5]Weiss LM, Dubey JP.Toxoplasmosis: A history of clinical observations[J]. Int J Parasitol, 2009, 39: 895-901.DOI: 10.1016/j.ijpara.2009.02.004
[6]Zhou P, Zhang H, Lin RQ, et al. Genetic characterization ofToxoplasmagondiiisolates from China[J]. Parasitol Int, 2009, 58: 193-195.DOI: 10.1016/j.parint.2009.01.006
[7]Kijlstra A, Jongert E.Toxoplasma-safe meat: close to reality[J]. Trends Parasitol,2009, 25: 18-22.DOI: 10.1016/j.pt.2008.09.008
[8]Innes EA.A brief history and overview ofToxoplasmagondii[J]. Zoonoses Public Hlth, 2010, 57: 1-7.DOI: 10.1111/j.1863-2378.2009.01276.x
[9]Robert GF, Darde ML. Epidemiology of and diagnostic strategies for toxoplasmosis[J]. Clin Microbiol Rev, 2012, 25: 264-296.DOI:10.1128/CMR.05013-11
[10]Sibley LD, Ajioka JW. Population structure ofToxoplasmagondii: clonal expansion driven by infrequent recombination and selective sweeps[J]. Annu Rev Microbiol, 2008, 62: 329-351.DOI:10.1146/annurev.micro.62.081307.162925
[11]Brydges SD, Sherman GD, Nockemann S,et al.Molecular characterization of TgMIC5, a proteolytically processed antigen secreted from the micronemes ofToxoplasmagondii[J]. Mol Biochem Parasitol, 2000, 111:51-66.DOI:10.1016/S0166-6851(00)00296-6
[12]Brydges SD, Zhou XW,Huynh MH, et al. Targeted deletion ofMIC5 enhances trimming proteolysis ofToxoplasmainvasion proteins[J].Eukaryot Cell, 2006, 5: 2174-2183.DOI: 10.1128/EC.00163-06
[13]Su C, Shwab EK, Zhou P, et al.Moving towards an integrated approach to molecular detection and identification ofToxoplasmagondii[J]. Parasitology, 2010, 137: 1-11.DOI: 10.1017/S0031182009991065
[14]Zhou P, Nie H, Zhang LX, et al. Genetic characterization ofToxoplasmagondiiisolates from pigs in China[J]. J Parasitol, 2010, 96: 1027-1029.DOI:10.1645/GE-2465.1
[15]Xu Y, Zhang NZ, Chen J, et al.Toxoplasmagondiirhoptry protein 38 gene: sequence variation among isolates from different hosts and geographical locations[J]. Genet Mol Res, 2014, 13:4839-4844.DOI:10.4238/2014.January.14.3
[16]Chen J, Fang SF, Zhou DH, et al. Sequence variation in theToxoplasmagondiieIF4A gene among strains from different hosts and geographical locations[J]. Genet Mol Res, 2014, 13: 3356-3361.DOI:10.4238/2014.April.29.14
[17]Thompson JD, Gibson TJ, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acids Res, 1997, 25: 4876-4882.DOI:10.1093/nar/25.24.4876
[18]Ronquist F, Huelsenbeck JP.mrBayes 3: Bayesian phylogenetic inference under mixed models[J]. Bioinformatics, 2003, 19: 1572-1574.DOI:10.1093/bioinformatics/btg180
[19]Swofford DL.PAUP*: phylogenetic analysis using parsimony, version 4.0b10[P]. Sinauer Associates, Sunderland, MA 2002.
[20]Chen J, Li ZY, Zhou DH, et al. Genetic diversity amongToxoplasmagondiistrains from different hosts and geographical regions revealed by sequence analysis of GRA5 gene[J]. Parasit Vector, 2012, 5: 279.DOI: 10.1186/1756-3305-5-279
[21]Chen J, Fang SF, Li ZY, et al. Sequence variation in ROP9 gene amongToxoplasmagondiistrains from different hosts and geographical locations[J]. J Anim Vet Adv,2012, 11: 4288-4291.DOI: 10.3923/javaa.2012.4288.4291
[22]RenD, Zhou DH, Xu MJ, et al.Sequence variation inToxoplasmagondiiMIC13 gene among isolates from different hosts and geographical locations[J]. Afr J Microbiol Res, 2012,6: 1333-1337.
[23]Yan HK, Song HQ, Zhou Y, et al.Sequence variation in perforin-like protein 1 gene among sixToxoplasmagondiistrains[J]. J Anim Vet Adv, 2011, 10: 2244-2247.DOI:10.3923/javaa.2011.2244.2247
Received:2015-07-20;Revision accepted:2015-09-21
Sequence variability of MIC5 gene among 13Toxoplasmagondiiisolates belonging to different genotypes
LIU Wen-ge1,2,XU Xiao-pei1,2,CHEN Jia2,ZHU Xing-quan2,XU Qian-ming1
(1.CollegeofAnimalScienceandTechnology,AnhuiAgriculturalUniversity,Hefei230036,China;2.StateKeyLaboratoryofVeterinaryEtiologicalBiology,KeyLaboratoryofVeterinaryParasitologyofGansuProvince,LanzhouVeterinaryResearchInstitute,ChineseAcademyofAgriculturalSciences,Lanzhou730046,China)
Toxoplasmagondii(T.gondii) has the ability to infect almost all warm-blooded animals including humans, causing a serious toxoplasmosis. MIC5, a member of the microneme proteins, directly regulates the activity of proteases, which is associated with the proteolytic susceptibility of other microneme protein substrates. In this study, we examined the sequence variability of MIC5 gene among 13T.gondiiisolates belonging to different genotypes, which were collected from different hosts and geographical regions. Also, phylogenetic relationships among the examinedT.gondiiisolates were re-constructed using maximum parsimony (MP),maximum likelihood (ML) and Bayesian inference (BI) methods. The MIC5 gene from all the isolates possesses 1 975 bp in length. Sequence comparison revealed 52 variable nucleotide positions (0-1.3%), including 38 variations in 3 introns, and 14 variations in 4 exons, and their A+T contents were 51.70%-51.95%. Phylogenetic analysis showed that MIC5 gene is not an ideal marker for studying population genetics ofT.gondiidue to its low sequence diversity.
Toxoplasmagondii;MIC5;sequence variation;phylogenetic analyses
Xu Qian-ming,email:vetqmxu@163.com
徐民俊,Email:xuminjun@caas.cn
1.中國農業科學院蘭州獸醫研究所,家畜疫病病原生物學國家重點實驗室,蘭州 730046; 2.華南農業大學動物科學院,廣州 510642 3.揚州大學獸醫學院,揚州 225009
10.3969/j.issn.1002-2694.2015.11.011
R383
A
1002-2694(2015)11-1037-05
甘肅省創新研究群體計劃項目 (No.1210RJIA006)資助

Supported by grant from the National Natural Science Foundation of China (Grant No. 31228022) and the Science Fund for Creative Research Groups of Gansu Province (Grant No. 1210RJIA006)

