晉南牛與部分地方黃牛之間遺傳多樣性分析
王 曦,張元慶*,賀東昌,張喜忠,李 博,王棟才,靳 光,李福星,楊效民,徐 芳
(1.山西省農業科學院畜牧獸醫研究所,太原 030032; 2.山西省運城市黃牛場,運城 044500)
晉南牛與部分地方黃牛之間遺傳多樣性分析
王 曦1,張元慶1*,賀東昌1,張喜忠1,李 博1,王棟才1,靳 光1,李福星2,楊效民1,徐 芳1
(1.山西省農業科學院畜牧獸醫研究所,太原 030032; 2.山西省運城市黃牛場,運城 044500)
旨在利用微衛星技術檢測晉南牛、郟縣紅牛、魯西牛和秦川牛四大地方黃牛的牛群遺傳多樣樣及遺傳結構。采用16個微衛星DNA標記對4個牛群進行檢測分析。16個微衛星DNA標記中,除HEL9位點在所有牛群中呈單態外,其他15個位點的有效等位基因數為2~8個,平均有效等位基因數為3.067 6個。BM1818為低度多態(PIC=0.083 0,PIC<0.25),其余位點均為高度多態(PIC>0.5),其中HUAT24多態性最大(PIC=0.727 5)。4個牛群共檢測到80個等位基因,其中晉南牛為63個,郟縣紅牛為65個,魯西牛65個,秦川牛68個。在IDVGA46位點,僅有晉南牛有B等位基因,而在TGLA44位點,僅有晉南牛沒有B等位基因。4個牛群的平均表觀雜合度為0.385 2,平均期望雜合度為0.640 3,平均雜合度為0.578 0。晉南牛在NJ樹上單獨聚為一支,與魯西黃牛、郟縣紅牛及秦川牛的遺傳距離分別為0.809 3、0.759 8、0.807 1。結果表明,晉南牛遺傳資源獨特,群體遺傳多樣性豐富,是一個生長進化上較為封閉的群體。
微衛星;遺傳多樣性;晉南牛
畜禽品種是畜牧業生產的基礎,地方畜禽品種更是畜牧業的稀缺財富。我國養牛歷史悠久,不僅積累了豐富的飼養管理經驗,更選育出不少優良地方品種,其中,晉南牛、秦川牛、魯西黃牛與郟縣紅牛等地方品種均為我國著名的黃牛品種。20世紀70年代以來,在國外肉牛品種與市場經濟的沖擊下,我國地方牛品種群體不斷萎縮,質量下降甚至瀕危滅絕,遺傳資源遭受重大損失。
微衛星DNA標記具有多態性強,呈孟德爾共顯性遺傳,分布廣泛等特征,常用作動物遺傳多樣性評價、群體遺傳關系和分析生物系統進化等理想的遺傳標記[1-2]。本研究使用微衛星技術,以晉南牛、秦川牛、魯西黃牛和郟縣紅牛為研究對象,旨在闡明這幾個地方牛品種的群體遺傳結構和遺傳多樣性狀況,并進行對比,為有效地選種、選育及保種提供依據和指導。
1 材料與方法
1.1 材料
1.1.1 試驗對象 本試驗材料分別從保種場的晉南牛、郟縣紅牛、魯西黃牛和秦川牛保種場各取得耳組織樣品100份。
1.1.2 耳組織樣品的采集 用耳組織取樣鉗從牛耳前沿取樣,加入到裝有75%酒精的1.5 mL離心管中,置入冰壺中運回實驗室。
1.1.3 引物來源及微衛星位點選擇 微衛星位點應選取雜合度高、多態信息含量高、易擴增的位點。根據這些特點,在牛不同染色體上選16個多態性適中,擴增片段長度約200 bp的微衛星位點。本試驗所用的16對引物是從國際互聯網(http://sol.marc.usda.gov)檢索獲得,而后由華大基因合成。引物信息如表1所示。
1.1.4 PCR擴增 在200 μL EP管中加入:10×PCR緩沖液2.0 μL,2.5mmol·L-1dNTP 1.6 μL,TaqDNA聚合酶0.5 μL 1U,上下游引物各30 ng,基因組DNA約100 ng,加超純水至總反應體積20 μL。反應程序:94 ℃預變性5 min,35個循環(94 ℃變性30 s,55 ℃退火45 s,72 ℃延伸90 s),72 ℃延伸10 min,4 ℃保存。
1.2 方法
1.2.1 DNA提取及微衛星檢測 采用苯酚-氯仿抽提法進行基因組DNA提取,檢測其濃度、純度合格后,對基因組DNA適當稀釋,進行PCR擴增,經檢測PCR擴增產物為預計片段大小,再用聚丙烯酰胺凝膠電泳進行各微衛星位點的多態性檢測。
1.2.2 統計分析 等位基因數及各基因頻率,多態信息含量(Polymorphism information content,PIC),有效等位基因數,平均表觀雜合度,平均期望雜合度,平均雜合度等參數采用Microsatellite Toolkit軟件統計分析。群體間的Nei’sDA遺傳距離用Dispan軟件計算,并構建NJ樹。
2 結 果
2.1 基因組DNA及各微衛星位點產物凝膠檢測
將從牛耳組織中提取的全基因組DNA進行瓊脂糖凝膠(濃度為0.8%)電泳(圖1)。由圖1中可以看出,基因組DNA條帶清晰,點樣孔干凈,無拖尾現象,樣品亮度均一,無蛋白污染,不存在降解,可用于后續試驗。
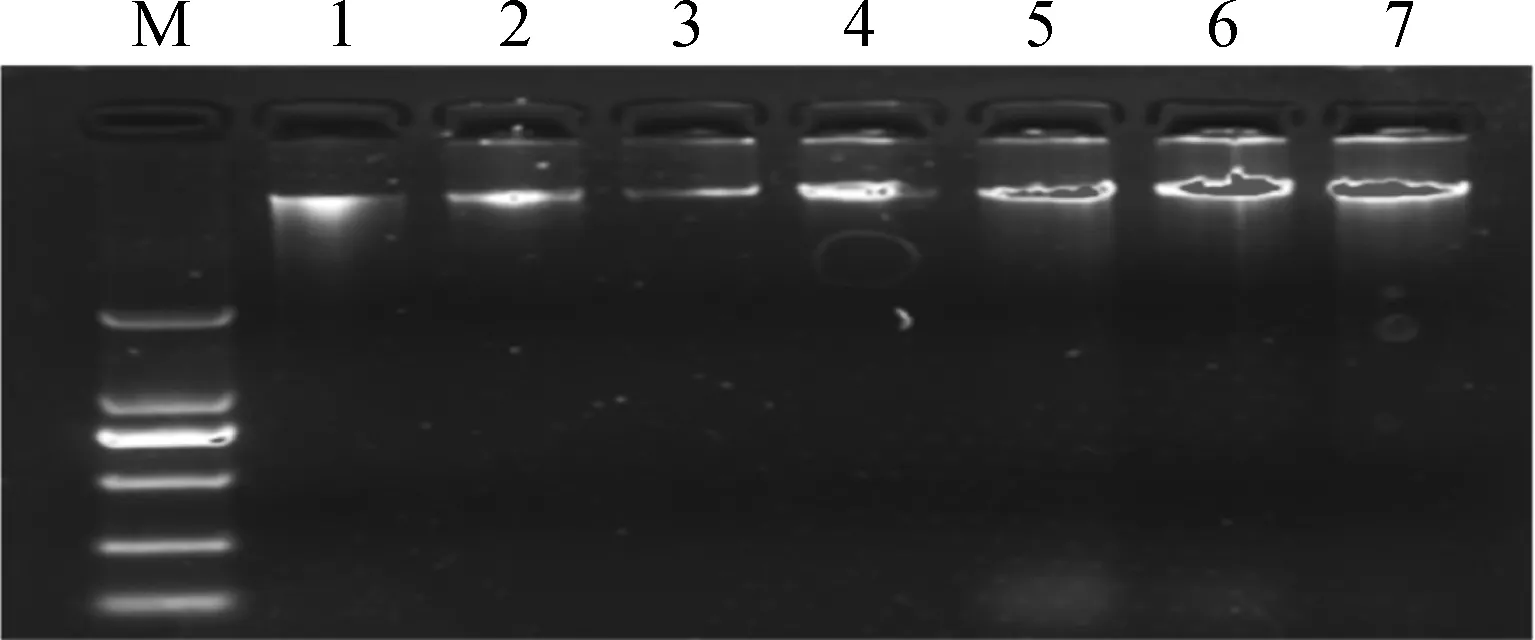
M.DNA相對分子質量標準DL2000;1~7.基因組DNAM.DL2000 marker;1-7.Genomic DNA 圖1 牛基因組DNA瓊脂糖凝膠檢測Fig.1 Agarose gel image of genomic DNA
微衛星PCR擴增產物瓊脂糖凝膠(濃度為1.5%)檢測結果如圖2所示,上樣PCR產物量為3 μL,DNA的量約為100 ng,結果顯示,PCR產物條帶整齊,特異性好,產物亮度表明PCR濃度較高,能夠滿足微衛星的基因型檢測。
微衛星PCR產物聚丙烯酰胺凝膠(濃度為8%)檢測(以微衛星位點ETH10為例)如圖3所示,樣品上樣量為6 μL,圖中雖然有影子帶存在,但是不影響基因型判定。
表1 微衛星位點、引物序列和退火溫度
Table 1 Microsatellite locus,sequences and annealing temperature
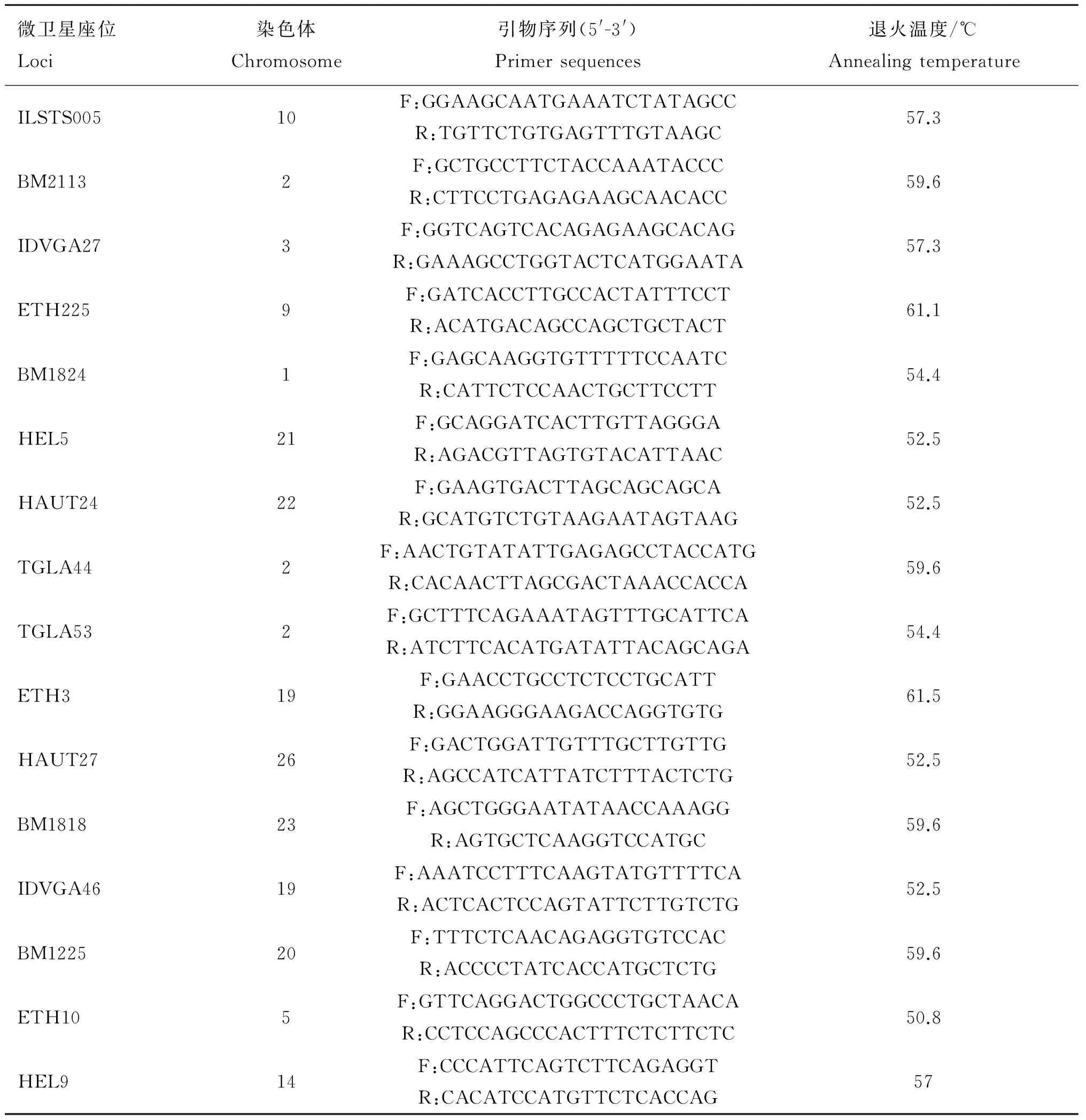
微衛星座位Loci染色體Chromosome引物序列(5'-3')Primersequences退火溫度/℃AnnealingtemperatureILSTS00510F:GGAAGCAATGAAATCTATAGCCR:TGTTCTGTGAGTTTGTAAGC57.3BM21132F:GCTGCCTTCTACCAAATACCCR:CTTCCTGAGAGAAGCAACACC59.6IDVGA273F:GGTCAGTCACAGAGAAGCACAGR:GAAAGCCTGGTACTCATGGAATA57.3ETH2259F:GATCACCTTGCCACTATTTCCTR:ACATGACAGCCAGCTGCTACT61.1BM18241F:GAGCAAGGTGTTTTTCCAATCR:CATTCTCCAACTGCTTCCTT54.4HEL521F:GCAGGATCACTTGTTAGGGAR:AGACGTTAGTGTACATTAAC52.5HAUT2422F:GAAGTGACTTAGCAGCAGCAR:GCATGTCTGTAAGAATAGTAAG52.5TGLA442F:AACTGTATATTGAGAGCCTACCATGR:CACAACTTAGCGACTAAACCACCA59.6TGLA532F:GCTTTCAGAAATAGTTTGCATTCAR:ATCTTCACATGATATTACAGCAGA54.4ETH319F:GAACCTGCCTCTCCTGCATTR:GGAAGGGAAGACCAGGTGTG61.5HAUT2726F:GACTGGATTGTTTGCTTGTTGR:AGCCATCATTATCTTTACTCTG52.5BM181823F:AGCTGGGAATATAACCAAAGGR:AGTGCTCAAGGTCCATGC59.6IDVGA4619F:AAATCCTTTCAAGTATGTTTTCAR:ACTCACTCCAGTATTCTTGTCTG52.5BM122520F:TTTCTCAACAGAGGTGTCCACR:ACCCCTATCACCATGCTCTG59.6ETH105F:GTTCAGGACTGGCCCTGCTAACAR:CCTCCAGCCCACTTTCTCTTCTC50.8HEL914F:CCCATTCAGTCTTCAGAGGTR:CACATCCATGTTCTCACCAG57
F.正向引物;R.反向引物
F.Forward primer;R.Reverse primer
2.2 各位點的群體研究
2.2.1 各等位基因及頻率 對晉南牛、郟縣紅牛、魯西牛和秦川牛群16個微衛星位點不同等位基因的檢測結果顯示,除HEL9為單態位點,其余位點均表現為多態,等位基因及頻率如表2~6所示,4個牛群共檢測到80個等位基因,其中晉南牛為63個,郟縣紅牛為65個,魯西牛65個,秦川牛68個。表2~6列出基因頻率及等位基因。試驗中檢測出了在一些品種特有的等位基因,例如:在IDVGA46位點,僅有晉南牛有B等位基因;而在TGLA44位點,只有晉南牛沒有B等位基因;其中ETH10與BM1225等位基因最多,為8個,ILSTS005等位基因最少,為2個。等位基因的大小主要集中于150~200 bp之間,如表6所示,其中BM1818位點的等位基因片段最大為226~246 bp,IDVGA27位點的片段最小為95~118 bp。
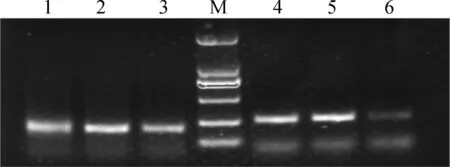
M.DNA相對分子質量標準DL2000;1~3.BM1225位點,PCR產物片段長度約為210 bp;4~6.BM1818位點,PCR產物片段長度約為250 bpM.DL2000 marker;1-3.PCR products of BM1225 locus about 210 bp;4-6.PCR products of BM1818 locus about 250 bp圖2 微衛星位點BM1225和BM1818 PCR產物瓊脂糖檢測Fig.2 Agarose gel image of detecting PCR products of BM1225 and BM1818 loci
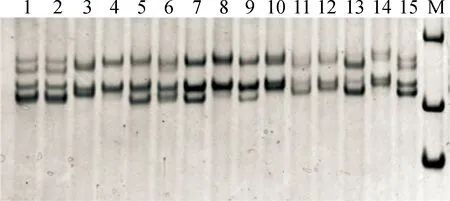
1,2,5,7.AD;3.BC;4,8.DD;6,9,13.AC;10,12,14.CD;11.BD;15.AB;M.Marker,from down to up were 100,200 and 300 bp,respectively圖3 微衛星位點ETH10 PCR產物聚丙烯酰胺凝膠電泳結果Fig.3 PAGE image of PCR product of ETH10 locus
表2 4個群體等位基因頻率
Table 2 Allele frequency of loci in 4 populations
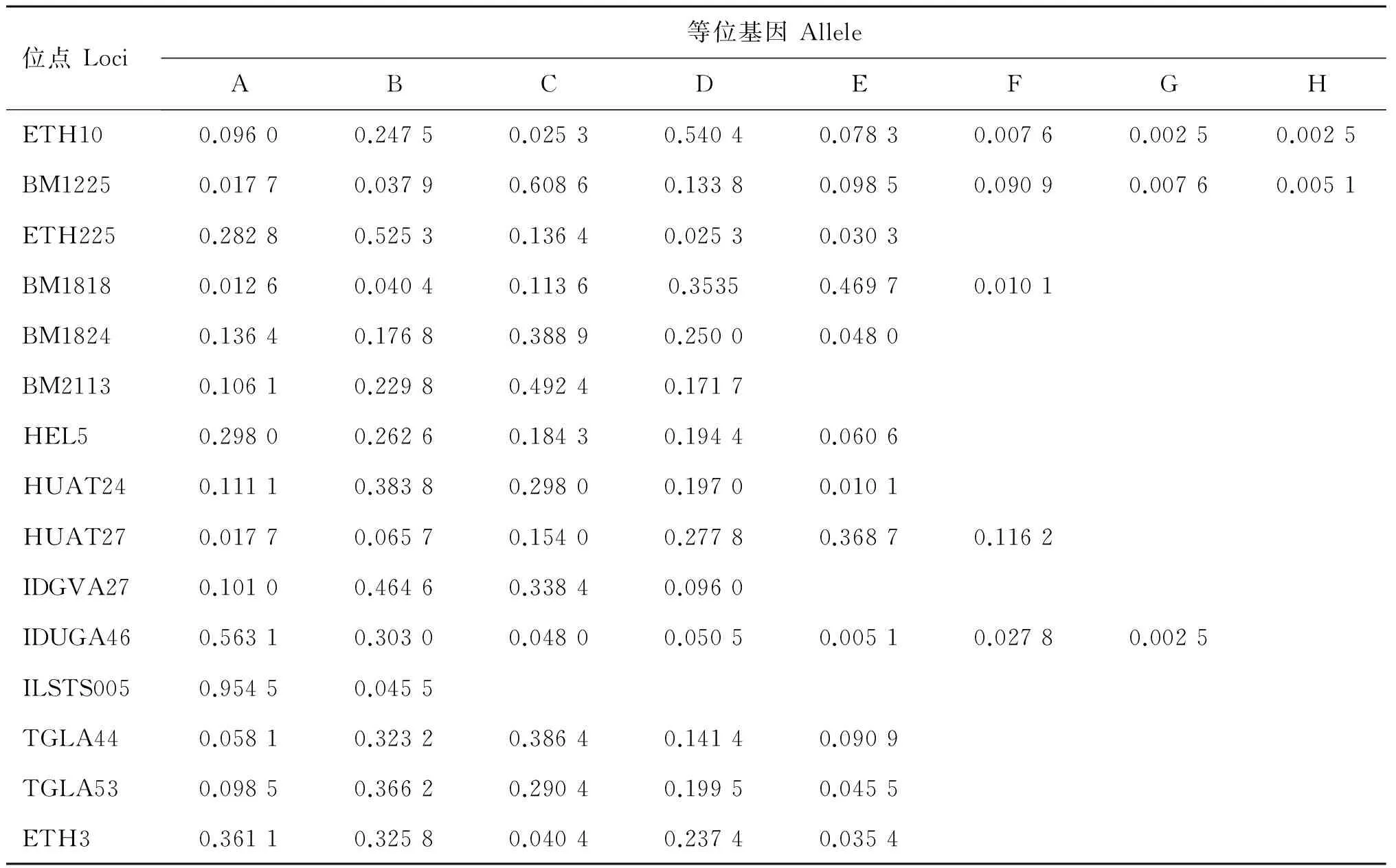
位點Loci等位基因AlleleABCDEFGHETH100.09600.24750.02530.54040.07830.00760.00250.0025BM12250.01770.03790.60860.13380.09850.09090.00760.0051ETH2250.28280.52530.13640.02530.0303BM18180.01260.04040.11360.35350.46970.0101BM18240.13640.17680.38890.25000.0480BM21130.10610.22980.49240.1717HEL50.29800.26260.18430.19440.0606HUAT240.11110.38380.29800.19700.0101HUAT270.01770.06570.15400.27780.36870.1162IDGVA270.10100.46460.33840.0960IDUGA460.56310.30300.04800.05050.00510.02780.0025ILSTS0050.95450.0455TGLA440.05810.32320.38640.14140.0909TGLA530.09850.36620.29040.19950.0455ETH30.36110.32580.04040.23740.0354
2.2.2 各微衛星位點遺傳特性分析 4個牛群中的多態信息含量結果如表7所示,BM1818為低度多態位點(PIC=0.083 0,PIC<0.25),其余位點均為高度多態(PIC>0.5),其中HUAT24多態信息含量值最大(PIC=0.727 5)。在晉南牛群中,BM1818為低度多態位點(PIC<0.25),BM2113、ETH、HEL5及TGLA44屬于中度多態位點(0.5>PIC>0.25),其余位點為高度多態位點(PIC>0.5)。郟縣紅牛群中,BM1818為低度多態位點,ETH225、ETH3及HUAT27為中度多態位點,其余均為高度多態位點。魯西黃牛中,BM1818為低度多態位點,BM1824為中度多態位點,其余為高度多態位點。秦川牛中,BM1818與ETH10為低度多態位點,BM2113、IDVGA27、ETH225及HUAT27等為中度多態位點,其余均為高度多態位點。
表3 晉南牛群體等位基因頻率
Table 3 Allele frequency of loci in Jinnan cattle
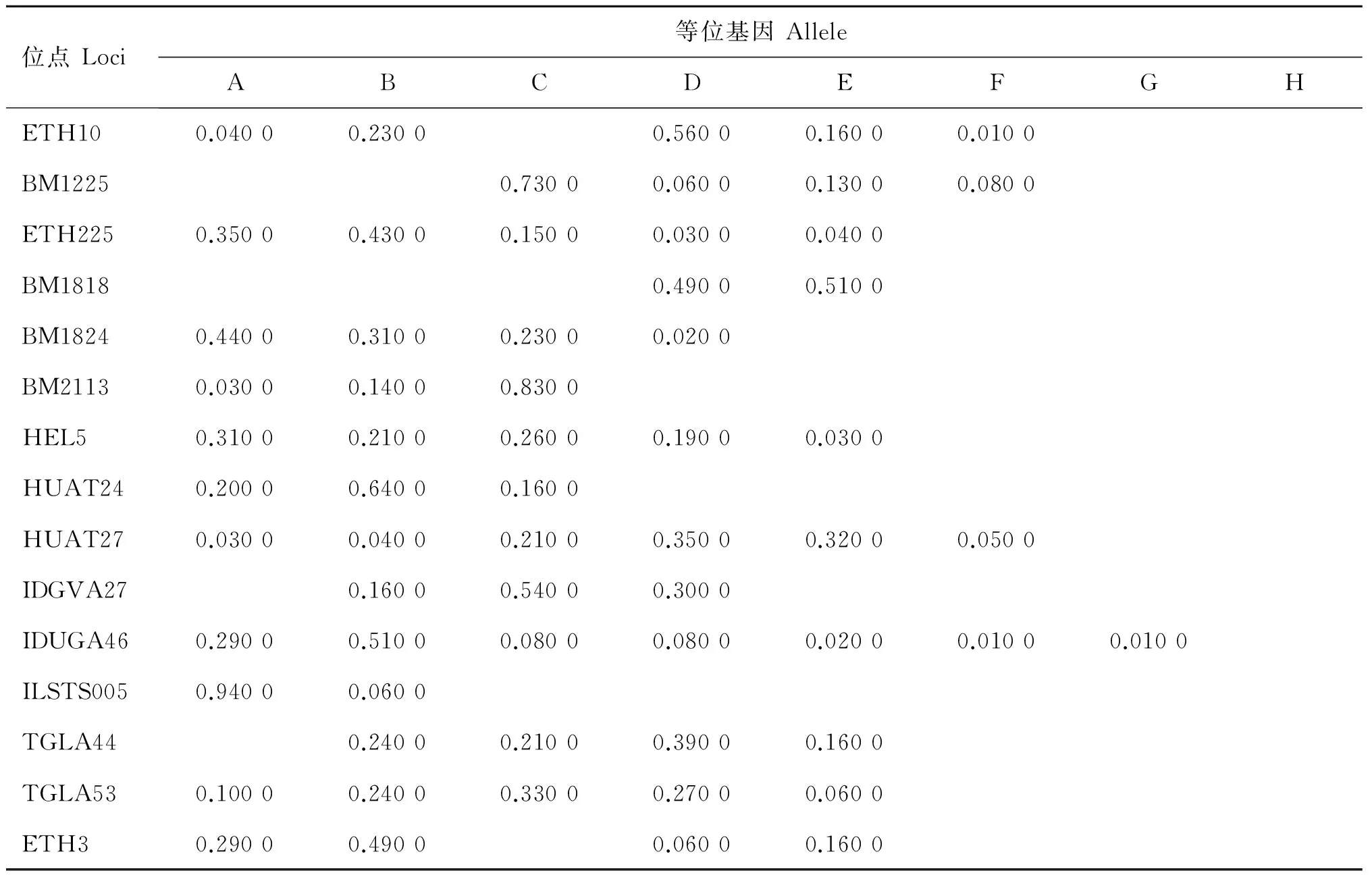
位點Loci等位基因AlleleABCDEFGHETH100.04000.23000.56000.16000.0100BM12250.73000.06000.13000.0800ETH2250.35000.43000.15000.03000.0400BM18180.49000.5100BM18240.44000.31000.23000.0200BM21130.03000.14000.8300HEL50.31000.21000.26000.19000.0300HUAT240.20000.64000.1600HUAT270.03000.04000.21000.35000.32000.0500IDGVA270.16000.54000.3000IDUGA460.29000.51000.08000.08000.02000.01000.0100ILSTS0050.94000.0600TGLA440.24000.21000.39000.1600TGLA530.10000.24000.33000.27000.0600ETH30.29000.49000.06000.1600
表4 郟縣紅牛群體等位基因頻率
Table 4 Allele frequency of loci in Jiaxian Red cattle
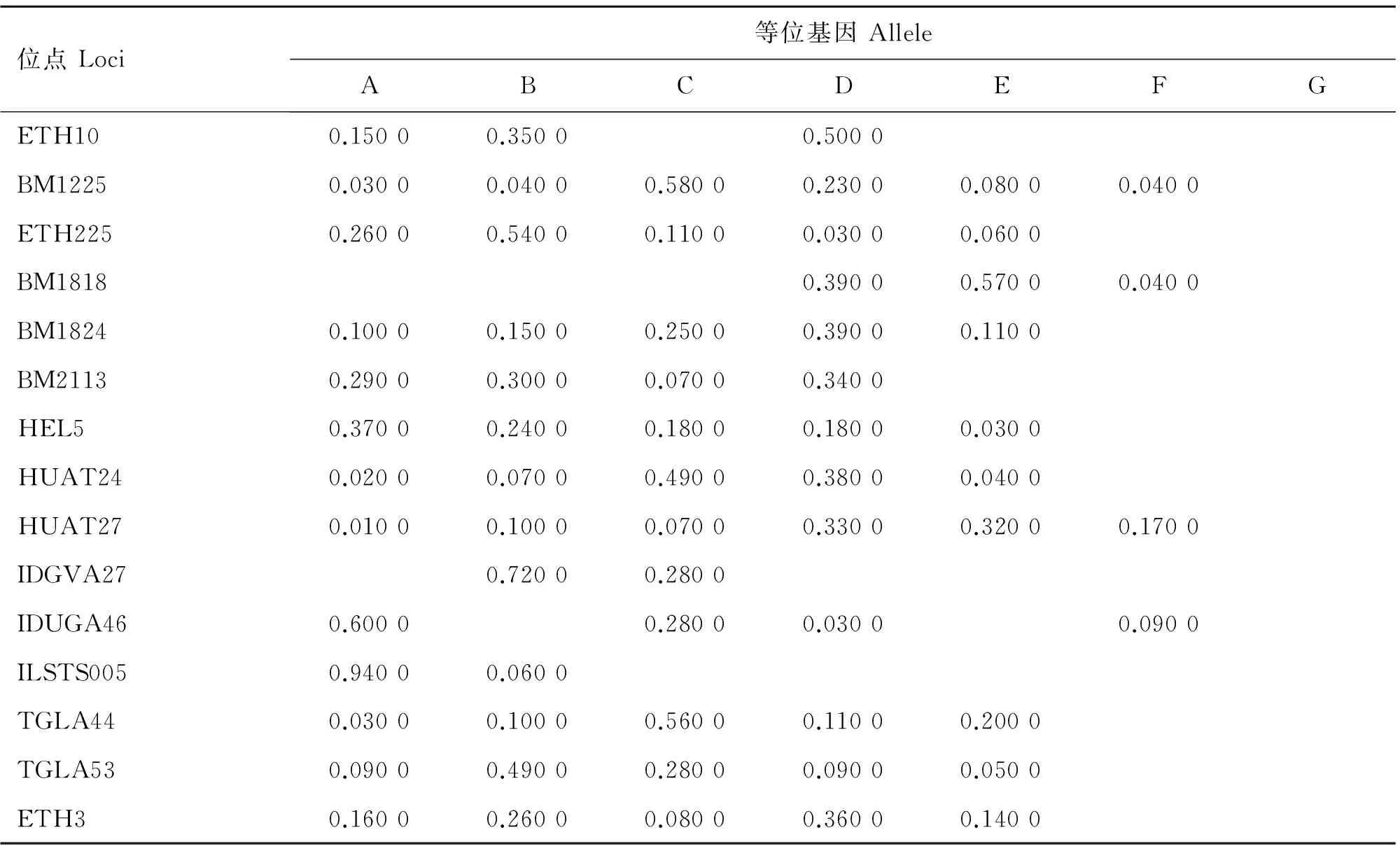
位點Loci等位基因AlleleABCDEFGETH100.15000.35000.5000BM12250.03000.04000.58000.23000.08000.0400ETH2250.26000.54000.11000.03000.0600BM18180.39000.57000.0400BM18240.10000.15000.25000.39000.1100BM21130.29000.30000.07000.3400HEL50.37000.24000.18000.18000.0300HUAT240.02000.07000.49000.38000.0400HUAT270.01000.10000.07000.33000.32000.1700IDGVA270.72000.2800IDUGA460.60000.28000.03000.0900ILSTS0050.94000.0600TGLA440.03000.10000.56000.11000.2000TGLA530.09000.49000.28000.09000.0500ETH30.16000.26000.08000.36000.1400
表5 魯西牛群體等位基因頻率
Table 5 Allele frequency of loci in Luxi cattle
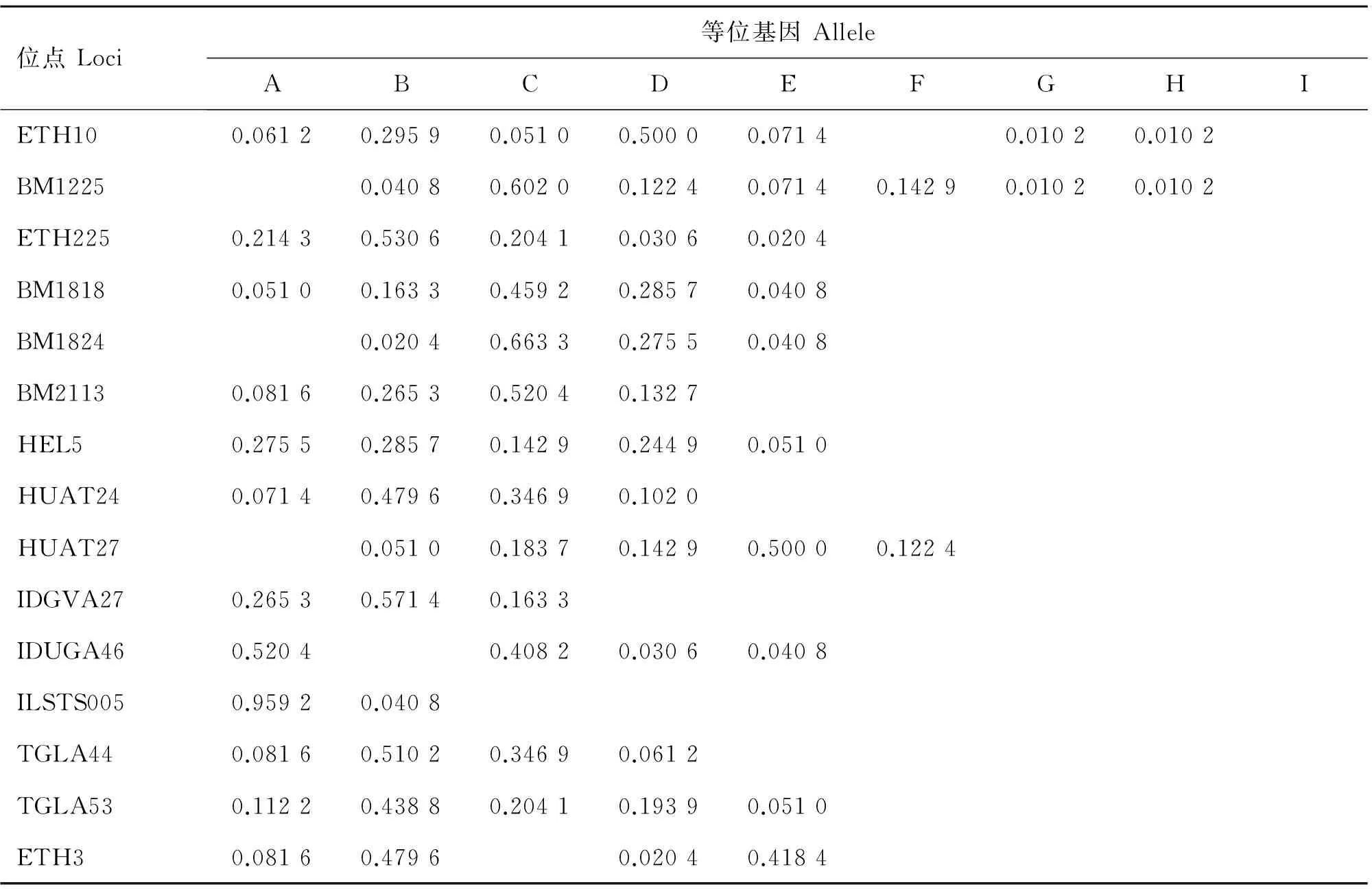
位點Loci等位基因AlleleABCDEFGHIETH100.06120.29590.05100.50000.07140.01020.0102BM12250.04080.60200.12240.07140.14290.01020.0102ETH2250.21430.53060.20410.03060.0204BM18180.05100.16330.45920.28570.0408BM18240.02040.66330.27550.0408BM21130.08160.26530.52040.1327HEL50.27550.28570.14290.24490.0510HUAT240.07140.47960.34690.1020HUAT270.05100.18370.14290.50000.1224IDGVA270.26530.57140.1633IDUGA460.52040.40820.03060.0408ILSTS0050.95920.0408TGLA440.08160.51020.34690.0612TGLA530.11220.43880.20410.19390.0510ETH30.08160.47960.02040.4184
表6 秦川牛群體等位基因頻率
Table 6 Allele frequency of loci in Qinchuan cattle
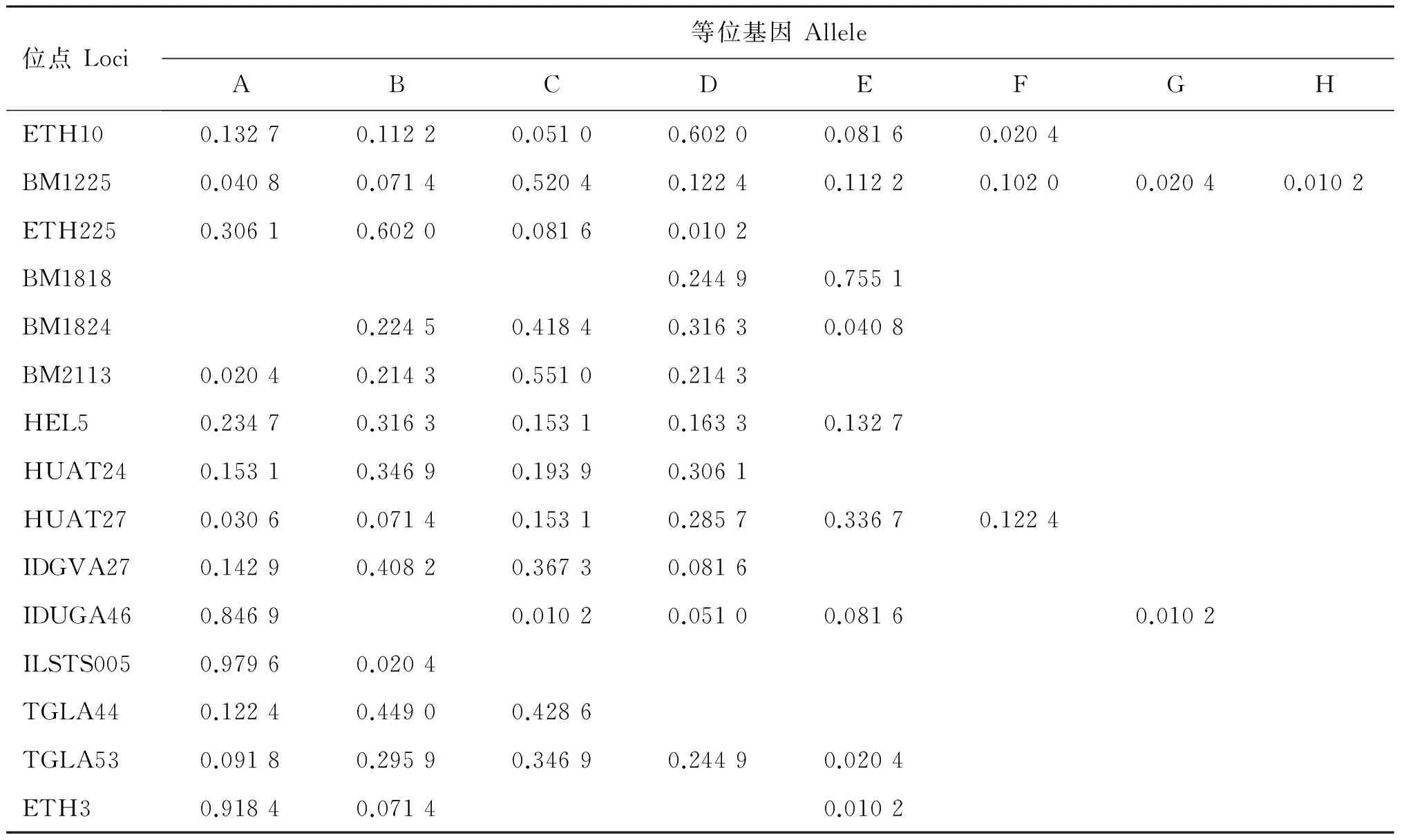
位點Loci等位基因AlleleABCDEFGHETH100.13270.11220.05100.60200.08160.0204BM12250.04080.07140.52040.12240.11220.10200.02040.0102ETH2250.30610.60200.08160.0102BM18180.24490.7551BM18240.22450.41840.31630.0408BM21130.02040.21430.55100.2143HEL50.23470.31630.15310.16330.1327HUAT240.15310.34690.19390.3061 HUAT270.03060.07140.15310.28570.33670.1224IDGVA270.14290.40820.36730.0816IDUGA460.84690.01020.05100.08160.0102ILSTS0050.97960.0204TGLA440.12240.44900.4286TGLA530.09180.29590.34690.24490.0204ETH30.91840.07140.0102
表7 4個群體中微衛星標記PCR擴增條件、片段大小、等位基因數及PIC值
Table 7PICvalue,length and the number of alleles of all SSRs loci in 4 populations
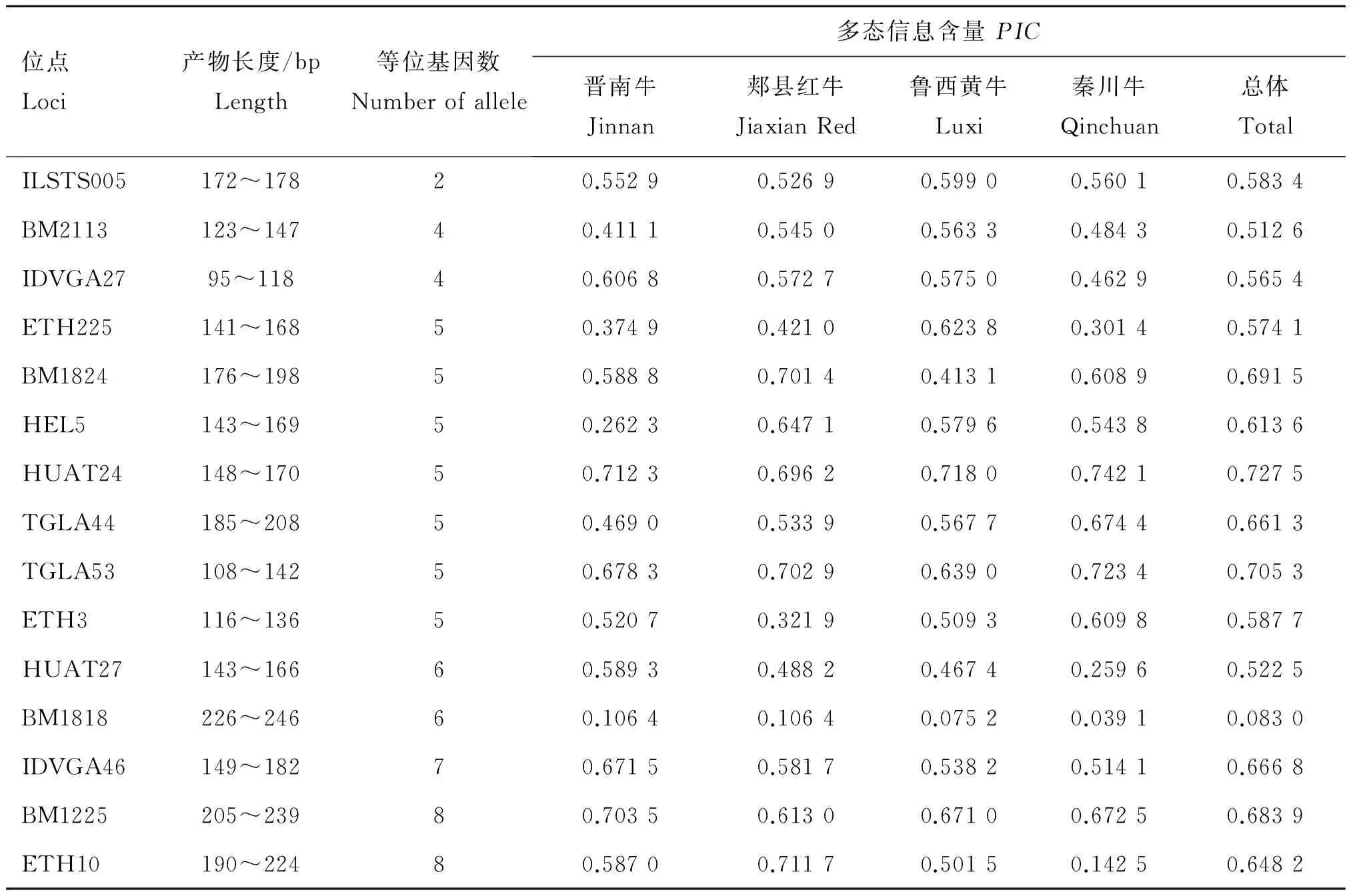
位點Loci產物長度/bpLength等位基因數Numberofallele多態信息含量PIC晉南牛Jinnan郟縣紅牛JiaxianRed魯西黃牛Luxi秦川牛Qinchuan總體TotalILSTS005172~17820.55290.52690.59900.56010.5834BM2113123~14740.41110.54500.56330.48430.5126IDVGA2795~11840.60680.57270.57500.46290.5654ETH225141~16850.37490.42100.62380.30140.5741BM1824176~19850.58880.70140.41310.60890.6915HEL5143~16950.26230.64710.57960.54380.6136HUAT24148~17050.71230.69620.71800.74210.7275TGLA44185~20850.46900.53390.56770.67440.6613TGLA53108~14250.67830.70290.63900.72340.7053ETH3116~13650.52070.32190.50930.60980.5877HUAT27143~16660.58930.48820.46740.25960.5225BM1818226~24660.10640.10640.07520.03910.0830IDVGA46149~18270.67150.58170.53820.51410.6668BM1225205~23980.70350.61300.67100.67250.6839ETH10190~22480.58700.71170.50150.14250.6482
由表8~12所示,本試驗所選15個微衛星多態位點在4個群體中平均有效等位基因數為3.067 6,晉南牛為2.682 5,郟縣紅牛為2.810 7,魯西黃牛為2.665 2,秦川牛為2.668 5。4個群體中平均純合度觀測值為0.614 8,晉南牛為0.622 7,郟縣紅牛為0.562 7,魯西黃牛為0.635 4,秦川牛為0.639 5。4個群體的平均雜合度觀測值為0.385 2,晉南牛為0.377 3,郟縣紅牛為0.562 7,魯西黃牛為0.364 6,秦川牛為0.360 5。4個群體中平均純合度期望值為0.359 7,晉南牛為0.418 9,郟縣紅牛為0.394 2,魯西黃牛為0.402 9,秦川牛為0.448 6。4個群體中平均雜合度期望值為0.640 3,晉南牛為0.581 1,郟縣紅牛為0.605 8,魯西黃牛為0.597 1,秦川牛為0.551 4。4個群體中平均Nei氏期望雜合度值為0.638 7,晉南牛為0.575 3,郟縣紅牛為0.599 7,魯西黃牛為0.591 0,秦川牛為0.545 8。4個群體中總平均雜合度值為0.578 0。
2.3 群體遺傳分化
H.Takahashi等[3]和M.Nei[4]通過計算機模擬對各種遺傳距離進行研究,證明在分析物種內群體間的遺傳變異時,運用DA遺傳距離是獲得準確系統發生樹的最有效的方法,且用UPGMA法優于NJ法。4個牛群之間的DA遺傳距離見表13,晉南牛與魯西黃牛、郟縣紅牛及秦川牛的遺傳距離分別為:0.809 3、0.759 8、0.807 1。基于4個牛群間DA遺傳距離構建的NJ遺傳關系樹,明顯地聚為3大支:第一支為晉南牛,第二支為秦川牛,第三支為郟縣紅牛和魯西牛(圖4)。

圖4 基于NJ法構建的4個牛群之間的遺傳關系樹Fig.4 Neighbour-joining (NJ) tree of 4 populations based on genetic distance
3 討 論
3.1 各等位基因與頻率
微衛星座位上等位基因數目和頻率的差異可以反映出家畜品種內和品種間的差異,近年來,使用微衛星DNA標記已經開展了一些地方牛群體遺傳特征和群體遺傳變異的工作。孫維斌等[5]利用TGLA227、ETH225、HEL9、CSSM66、TGLA1216等5個微衛星檢測晉南牛遺傳多態性,結果發現,位點均處于不平衡狀態(P<0.01),平均有效等位基因數為10.163 6。畢偉偉[6]利用8個微衛星研究了魯西牛群體的遺傳多態性,共檢測到34個等位基因,每個微衛星座位的等位基因數為3~6個,平均等位基因數為4.25個。王洪程[7]利用微衛星研究秦川牛群體的遺傳多態性,在144頭秦川牛的7個微衛星位點檢測到164個等位基因,平均每個位點23.43個等位基因,在IDVGA2位點上檢測到等位基因最多,為28個,在IDVGA27位點上檢測到等位基因最少,為16個。李榮嶺等[8]利用微衛星標記對12個中外牛品種群體遺傳結構的研究表明,郟縣紅牛的平均等位基因數最多(10.58),其他地方品種牛都在8.4以上,而外國引進品種的等位基因數較少(7~7.8)。本研究中,15個微衛星在4個群體中總共檢測到80個等位基因,其中晉南牛為63個,郟縣紅牛為65個,魯西牛65個,秦川牛68個,等位基因多態性比較豐富。研究中檢測發現在15個微衛星中,IDVGA46位點僅有晉南牛有B等位基因,為特有等位基因,而在TGLA44位點僅有晉南牛沒有B等位基因,為共有等位基因。郟縣紅牛在ETH10位點缺少E等位基因,同時在BM1818有F等位基因,在HUAT24存在E等位基因,在ETH位點檢測到C等位基因。這些特有等位基因,若在更多的品種中檢測驗證,則可作為代表品種的分子標記。
表8 4個群體微衛星位點遺傳參數統計
Table 8 Genetic parameter statistics of all SSRs loci in 4 populations
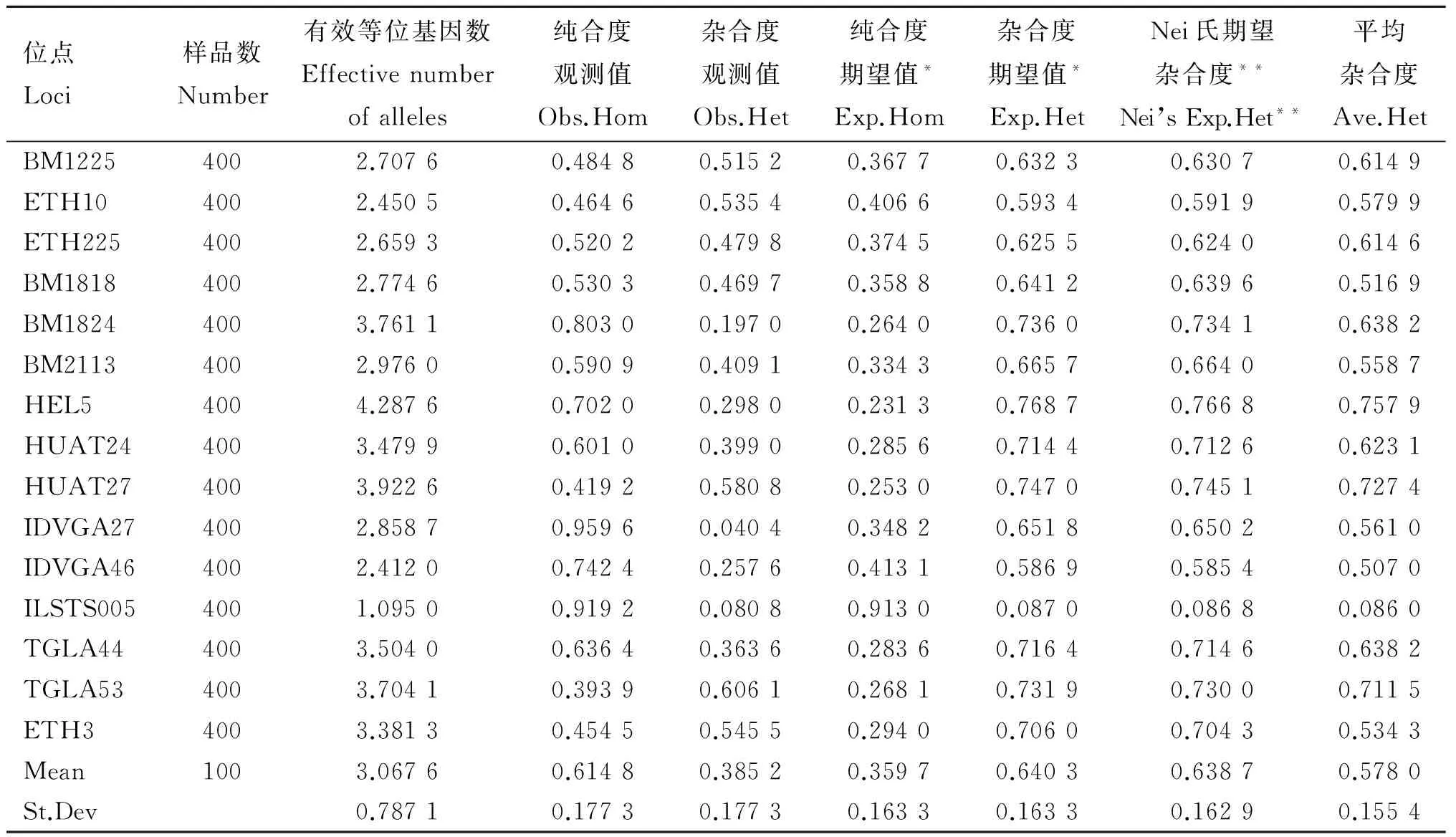
位點Loci樣品數Number有效等位基因數Effectivenumberofalleles純合度觀測值Obs.Hom雜合度觀測值Obs.Het純合度期望值*Exp.Hom雜合度期望值*Exp.HetNei氏期望雜合度**Nei’sExp.Het**平均雜合度Ave.HetBM12254002.70760.48480.51520.36770.63230.63070.6149ETH104002.45050.46460.53540.40660.59340.59190.5799ETH2254002.65930.52020.47980.37450.62550.62400.6146BM18184002.77460.53030.46970.35880.64120.63960.5169BM18244003.76110.80300.19700.26400.73600.73410.6382BM21134002.97600.59090.40910.33430.66570.66400.5587HEL54004.28760.70200.29800.23130.76870.76680.7579HUAT244003.47990.60100.39900.28560.71440.71260.6231HUAT274003.92260.41920.58080.25300.74700.74510.7274IDVGA274002.85870.95960.04040.34820.65180.65020.5610IDVGA464002.41200.74240.25760.41310.58690.58540.5070ILSTS0054001.09500.91920.08080.91300.08700.08680.0860TGLA444003.50400.63640.36360.28360.71640.71460.6382TGLA534003.70410.39390.60610.26810.73190.73000.7115ETH34003.38130.45450.54550.29400.70600.70430.5343Mean1003.06760.61480.38520.35970.64030.63870.5780St.Dev0.78710.17730.17730.16330.16330.16290.1554
*.期望雜合度和純合度由Levene計算而來;**.Nei’s期望雜合度。表9~12同
*.Expected homozygosty and heterozygosity were computed using Levene (1949);**.Nei’s (1973) expected heterozygosity.The same as Table 9-12
表9 晉南牛微衛星位點遺傳參數統計
Table 9 Genetic parameter statistics of all SSRs loci in Jinnan cattle
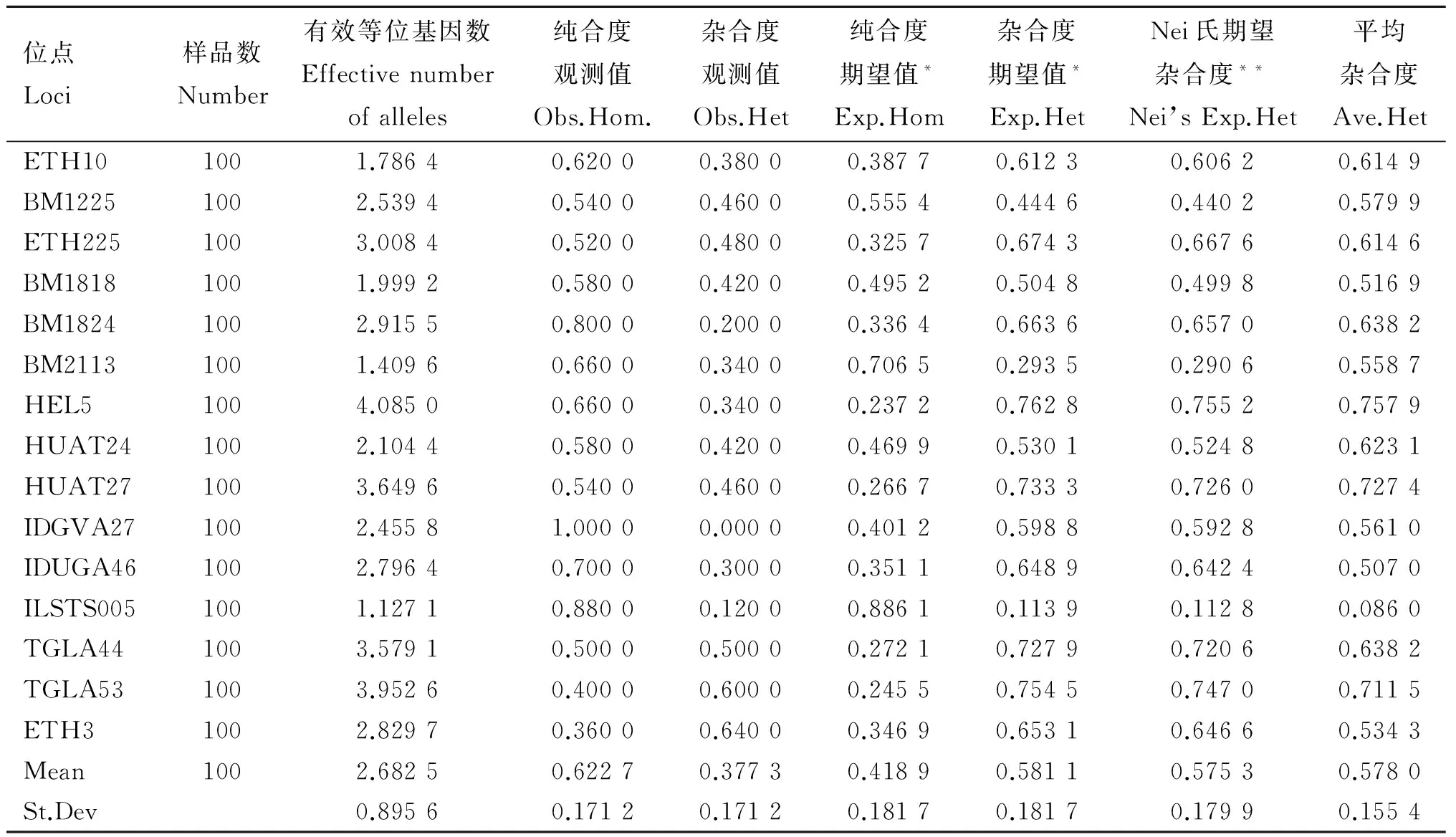
位點Loci樣品數Number有效等位基因數Effectivenumberofalleles純合度觀測值Obs.Hom.雜合度觀測值Obs.Het純合度期望值*Exp.Hom雜合度期望值*Exp.HetNei氏期望雜合度**Nei’sExp.Het平均雜合度Ave.HetETH101001.78640.62000.38000.38770.61230.60620.6149BM12251002.53940.54000.46000.55540.44460.44020.5799ETH2251003.00840.52000.48000.32570.67430.66760.6146BM18181001.99920.58000.42000.49520.50480.49980.5169BM18241002.91550.80000.20000.33640.66360.65700.6382BM21131001.40960.66000.34000.70650.29350.29060.5587HEL51004.08500.66000.34000.23720.76280.75520.7579HUAT241002.10440.58000.42000.46990.53010.52480.6231HUAT271003.64960.54000.46000.26670.73330.72600.7274IDGVA271002.45581.00000.00000.40120.59880.59280.5610IDUGA461002.79640.70000.30000.35110.64890.64240.5070ILSTS0051001.12710.88000.12000.88610.11390.11280.0860TGLA441003.57910.50000.50000.27210.72790.72060.6382TGLA531003.95260.40000.60000.24550.75450.74700.7115ETH31002.82970.36000.64000.34690.65310.64660.5343Mean1002.68250.62270.37730.41890.58110.57530.5780St.Dev0.89560.17120.17120.18170.18170.17990.1554
表10 郟縣紅牛微衛星位點遺傳參數統計
Table 10 Genetic parameter statistics of all SSRs loci in Jiaxian Red cattle
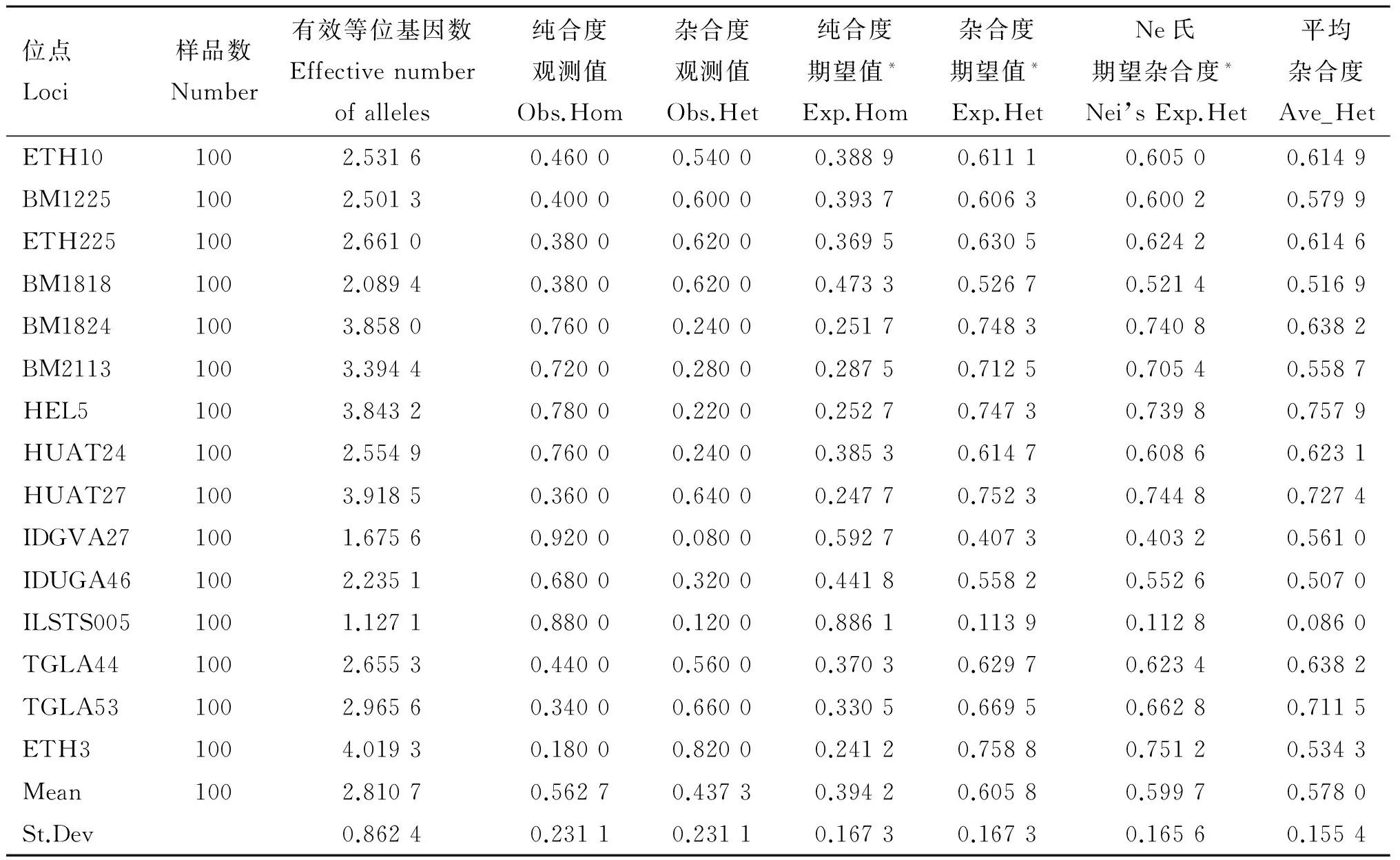
位點Loci樣品數Number有效等位基因數Effectivenumberofalleles純合度觀測值Obs.Hom雜合度觀測值Obs.Het純合度期望值*Exp.Hom雜合度期望值*Exp.HetNe氏期望雜合度*Nei’sExp.Het平均雜合度Ave_HetETH101002.53160.46000.54000.38890.61110.60500.6149BM12251002.50130.40000.60000.39370.60630.60020.5799ETH2251002.66100.38000.62000.36950.63050.62420.6146BM18181002.08940.38000.62000.47330.52670.52140.5169BM18241003.85800.76000.24000.25170.74830.74080.6382BM21131003.39440.72000.28000.28750.71250.70540.5587HEL51003.84320.78000.22000.25270.74730.73980.7579HUAT241002.55490.76000.24000.38530.61470.60860.6231HUAT271003.91850.36000.64000.24770.75230.74480.7274IDGVA271001.67560.92000.08000.59270.40730.40320.5610IDUGA461002.23510.68000.32000.44180.55820.55260.5070ILSTS0051001.12710.88000.12000.88610.11390.11280.0860TGLA441002.65530.44000.56000.37030.62970.62340.6382TGLA531002.96560.34000.66000.33050.66950.66280.7115ETH31004.01930.18000.82000.24120.75880.75120.5343Mean1002.81070.56270.43730.39420.60580.59970.5780St.Dev0.86240.23110.23110.16730.16730.16560.1554
表11 魯西黃牛微衛星位點遺傳參數
Table 11 Genetic parameter statistics of all SSRs loci in Luxi cattle
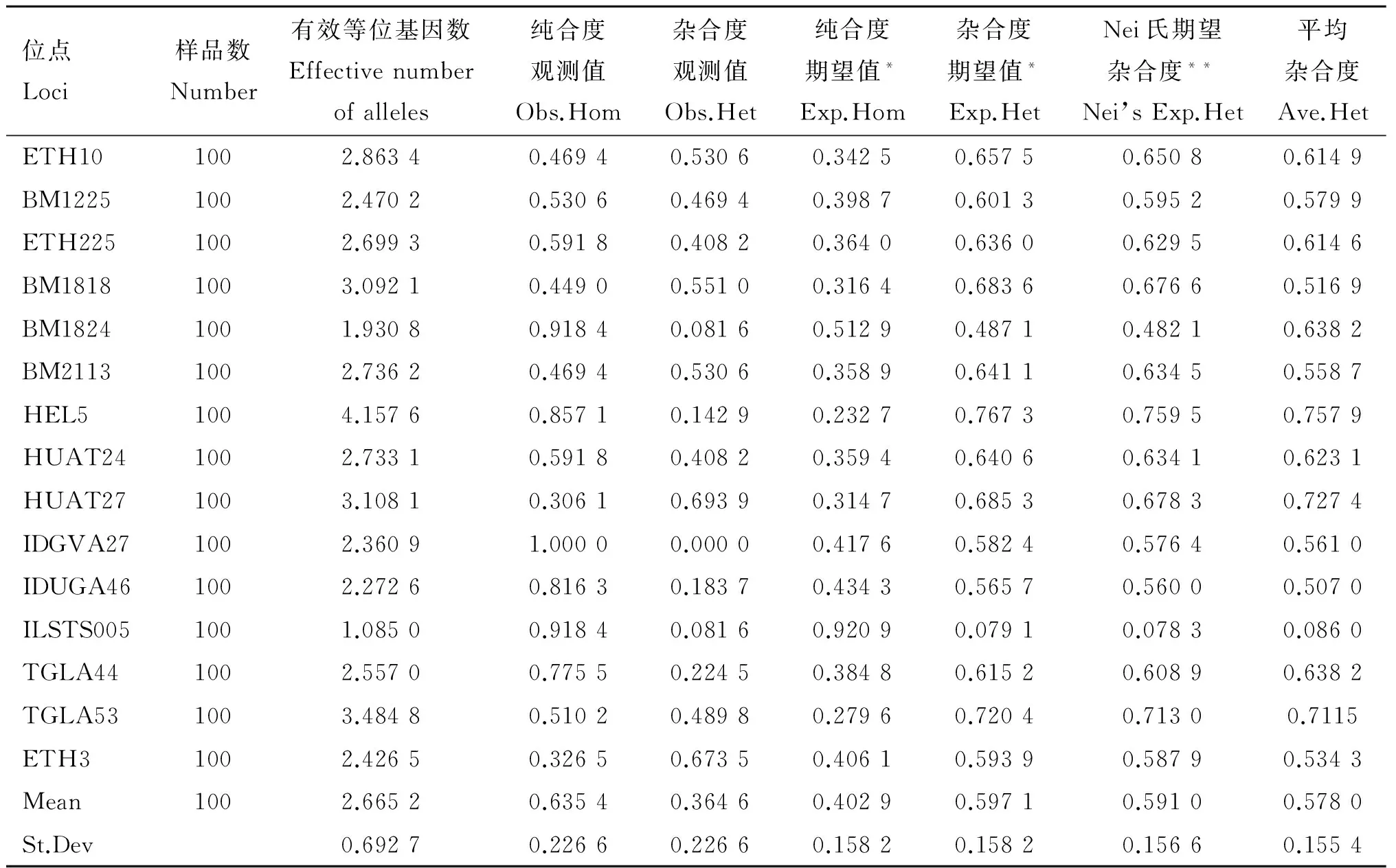
位點Loci樣品數Number有效等位基因數Effectivenumberofalleles純合度觀測值Obs.Hom雜合度觀測值Obs.Het純合度期望值*Exp.Hom雜合度期望值*Exp.HetNei氏期望雜合度**Nei’sExp.Het平均雜合度Ave.HetETH101002.86340.46940.53060.34250.65750.65080.6149BM12251002.47020.53060.46940.39870.60130.59520.5799ETH2251002.69930.59180.40820.36400.63600.62950.6146BM18181003.09210.44900.55100.31640.68360.67660.5169BM18241001.93080.91840.08160.51290.48710.48210.6382BM21131002.73620.46940.53060.35890.64110.63450.5587HEL51004.15760.85710.14290.23270.76730.75950.7579HUAT241002.73310.59180.40820.35940.64060.63410.6231HUAT271003.10810.30610.69390.31470.68530.67830.7274IDGVA271002.36091.00000.00000.41760.58240.57640.5610IDUGA461002.27260.81630.18370.43430.56570.56000.5070ILSTS0051001.08500.91840.08160.92090.07910.07830.0860TGLA441002.55700.77550.22450.38480.61520.60890.6382TGLA531003.48480.51020.48980.27960.72040.71300.7115ETH31002.42650.32650.67350.40610.59390.58790.5343Mean1002.66520.63540.36460.40290.59710.59100.5780St.Dev0.69270.22660.22660.15820.15820.15660.1554
表12 秦川牛微衛星位點遺傳參數統計
Table 12 Genetic parameter statistics of all SSRs loci in Qincuan cattle
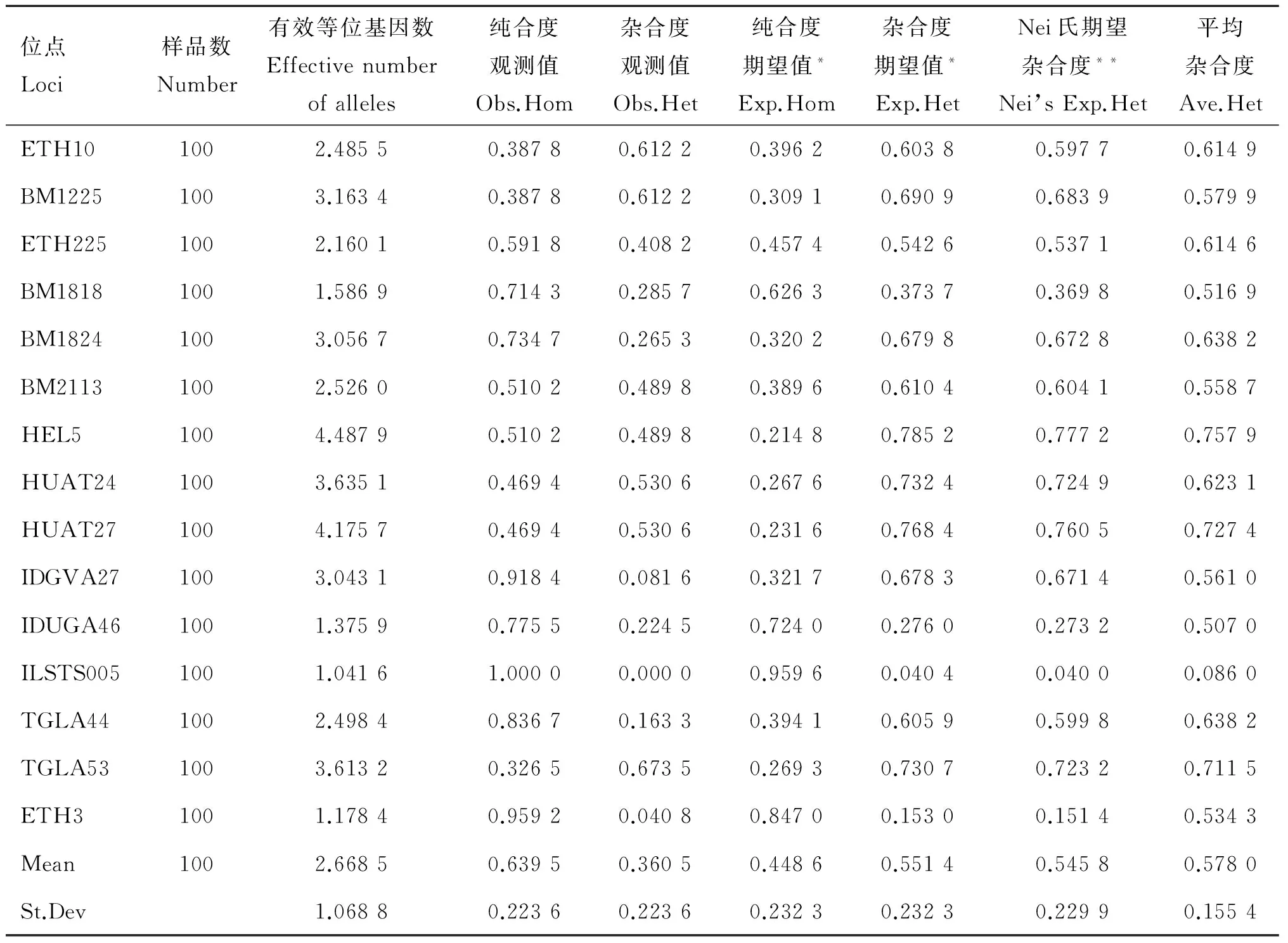
位點Loci樣品數Number有效等位基因數Effectivenumberofalleles純合度觀測值Obs.Hom雜合度觀測值Obs.Het純合度期望值*Exp.Hom雜合度期望值*Exp.HetNei氏期望雜合度**Nei’sExp.Het平均雜合度Ave.HetETH101002.48550.38780.61220.39620.60380.59770.6149BM12251003.16340.38780.61220.30910.69090.68390.5799ETH2251002.16010.59180.40820.45740.54260.53710.6146BM18181001.58690.71430.28570.62630.37370.36980.5169BM18241003.05670.73470.26530.32020.67980.67280.6382BM21131002.52600.51020.48980.38960.61040.60410.5587HEL51004.48790.51020.48980.21480.78520.77720.7579HUAT241003.63510.46940.53060.26760.73240.72490.6231HUAT271004.17570.46940.53060.23160.76840.76050.7274IDGVA271003.04310.91840.08160.32170.67830.67140.5610IDUGA461001.37590.77550.22450.72400.27600.27320.5070ILSTS0051001.04161.00000.00000.95960.04040.04000.0860TGLA441002.49840.83670.16330.39410.60590.59980.6382TGLA531003.61320.32650.67350.26930.73070.72320.7115ETH31001.17840.95920.04080.84700.15300.15140.5343Mean1002.66850.63950.36050.44860.55140.54580.5780St.Dev1.06880.22360.22360.23230.23230.22990.1554
表13 4個牛品種間Nei’s 遺傳距離(DA,對角線上方)和Nei's 標準遺傳距離(DS,對角線下方)
Table 13 Nei’s genetic distance (above the diagonal) and Nei's standard genetic distance (below the diagonal) among 4 indigenous cattle breeds
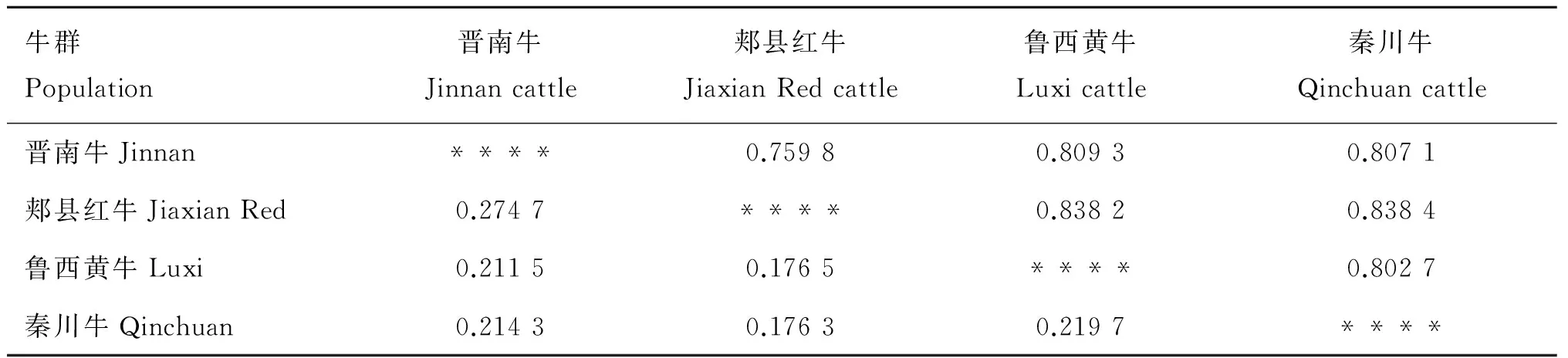
牛群Population晉南牛Jinnancattle郟縣紅牛JiaxianRedcattle魯西黃牛Luxicattle秦川牛Qinchuancattle晉南牛Jinnan****0.75980.80930.8071郟縣紅牛JiaxianRed0.2747****0.83820.8384魯西黃牛Luxi0.21150.1765****0.8027秦川牛Qinchuan0.21430.17630.2197****
本研究所檢測的微衛星位點中,等位基因數的范圍是2~7個,由于群體的規模有限,所測的等位基因數目與文獻報道有些差異,其中HEL9座位沒有檢測到多態,這與羅永生等的結果不同[9],ILSTS005位點等位基因數最少,僅檢測到2個,ETH10和BM1225位點等位基因數最多,共檢測到8個。也反映了微衛星位點在種間、群體間的差異,體現了其保守性。
從各微衛星的基因頻率來看,等位基因分布并不均勻,每個位點中都有一個或幾個優勢等位基因存在,且群體中頻率最高的等位基因是該物種中最為保守的,最為原始的,其他的等位基因則是由于該基因位點的突變引起的[10]。
3.2 位點遺傳特性分析
多態信息含量(PIC)是衡量標記多態性較好的指標,D.Bostein等[11]提出了衡量基因變異程度高低的多態信息指標:當PIC>0.5時,為高度多態性;當0.25 孫維斌等[5]檢測晉南牛TGLA1126位點雜合度、遺傳距離及PIC值,分別為0.976 5、8.940 7、0.885 8。5個微衛星平均雜合度為0.980 9,平均多態信息含量為0.904 9。王洪程等[7]利用微衛星研究秦川牛群體的遺傳多態性,7個微衛星位點在秦川牛群中表觀雜合度最低值與最高值分別為0.563 9和0.985 5,平均值為0.763 4,期望雜合度最低值與最高值分別為0.886 5和0.951 2,其平均值為0.923 2。7個位點PIC>0.5,其平均值為0.914 1,均呈高度多態性。本研究中,4個群體的平均雜合度期望值為0.640 3,晉南牛為0.581 1,郟縣紅牛為0.605 8,魯西黃牛為0.597 1,秦川牛為0.551 4。4個群體的平均Nei氏期望雜合度值為0.638 7,晉南牛為0.575 3,郟縣紅牛為0.599 7,魯西黃牛為0.591 0,秦川牛為0.545 8。4個群體的總平均雜合度值為0.578 0。所以期望的多樣性要高于觀測的多樣性,因此這4個種群可能存在選擇與近交[12]。 多態信息含量、有效等位基因數等都是衡量群體內遺傳變異大小的指標,這些數值在大小上均有一致性,多態信息含量大,有效等位基因數相對大[13-14]。這幾個參數較大時說明群體在該位點的變異性高,有較大的選擇潛力,即可利用該位點進行與生產性狀相關的標記輔助選擇[15]。 3.3 群體遺傳分化 晉南牛與郟縣紅牛、魯西黃牛、秦川牛的遺傳距離DA分別為0.759 8、0.809 3和0.807 1,距離值不太遠,可能是因為這4個品種在發展進化的過程中由于在地域上的封閉性及差異造成。晉南牛相對與其他3種牛比較獨立,可能由于晉南牛在選育過程中較為封閉,與其他品種的牛血緣上還沒有過多的交叉,具有典型的地域性,同時,由于郟縣紅牛與魯西黃牛在近年來突破了地域的局限性,基因相互有些滲透,所以同屬于共同的進化分支。M.A.Cronin 等利用微衛星標記研究美國平原野牛與森林野牛,結果表明,它們在群體遺傳結構上存在有更多的同源性[16]。T.Keros等利用微衛星對克羅地亞本地灰牛群分類后在遺傳結構上與美洲牛群比較,證明其更傾向于歐洲的牛群[17]。 本研究對晉南牛的遺傳結構及遺傳分化做了分子水平的研究,晉南牛在分子遺傳關系樹上獨成一支,這與雷初朝等利用mtDNA研究牛的分類所得出結論相一致[18]。這種群體遺傳特性上的獨立性,從分子水平上證明了晉南牛為我國寶貴的遺傳資源,因此對其的保種顯得尤為重要。利用微衛星檢測遺傳結構,可指導牛群的分類及保種工作。L.D.Pham等利用微衛星標記研究越南地方牛的遺傳多樣性,對6個類群從遺傳結構上進行分類分群,從而指導保種,大大節約了保種的費用,提高了效率[19]。Y.T.Utsunomiya 等利用AFLP遺傳標記研究西亞與南亞的瘤牛、水牛及已經滅絕的歐洲野牛牛群的遺傳結構,對牛群的保種起到積極的指導作用[20]。C.Acosta等對古巴5個牛群的遺傳結構利用微衛星進行了研究,結果表明,5個牛群可以接近于分成兩個牛群,對于牛群的保種有重要意義[21]。E.Armstrong等利用微衛星將烏拉圭牛與美洲牛群作對比,結果表明,遺傳漂變等可能會引起群體的遺傳變異,最終使得烏拉圭牛有別于美洲牛的遺傳群體,因此對其的保種有著非常重要的意義[22]。 [1] 張云武,張亞平.微衛星及其應用[J].動物學研究,2011,22(4):315-320. ZHANG Y W,ZHANG Y P.Microsatellites and applications [J].ZoologicalResearch,2011,22(4):315-320.(in Chinese) [2] BRUORD M W,BRADLEY D G,LUIKART G D N.A marker reveals the complexity of livestock domestication [J].NatRevGenet,2003,4(11):900-910. [3] TAKAHASHI H,NIRASAWA K,NAGAMINE Y,et al.Genetic relationship among Japanese native breeds of chicken based on microsatellite DNA polymorphisms[J].JHeredity,1998,89(6):543-546. [4] NEI M.Estimation of average heterozygosity and genetic distance from a small number of individual[J].Genetics,1978,89:583-590. [5] 孫維斌,陳 宏,樸亨國,等.晉南牛5個微衛星衛點多態性的研究[C].楊凌:全國首屆動物生物技術學術研討會,2004:345-348. SUN W B,CHENG H,PU H G,et al.Analysis of polymorphism of five microsatellites in Jin-nan cattle [C].Yangling:Animal Biotechnology Bulletin,2004:345-348.(in Chinese) [6] 畢偉偉.魯西牛微衛星標記和MHC多態性及其生長發育性狀的關系[D].泰安:山東農業大學,2011. BI W W.Polymorphism of microsatellite markers and MHC and their relationships with growth traits in Lu-xi cattle[D].Taian:Shandong Agricultural University,2011.(in Chinese) [7] 王洪程.秦川牛微衛星、CDIPT基因多態及其生長性狀的相關研究[D].楊凌:西北農林科技大學,2011. WANG H C.Polymorphism of microsatellites,CDIPT gene and their correlations with growth traits in Qinchuan cattle[D].Yangling:Northwest A&F University,2011.(in Chinese) [8] 李榮嶺,張桂香,王志剛,等.微衛星標記對12個中外牛品種群體遺傳結構的研究[J].遺傳,2007,29(12):1463-1470. LI R L,ZHANG G X,WANG Z G,et al.Analysis of the genetic structure of 12 Chinese and foreign cattle breeds using DNA microsatellite markers[J].Hereditas,2001,29(12):1463-1470.(in Chinese) [9] 羅永生,王志剛,李加琪,等.采用微衛星標記分析13個中外牛品種的遺傳變異和品種間的遺傳關系[J].生物多樣性,2006,14(6):498-507. LUO Y S,WANG Z G,LI J Q,et al.Genetic variation and genetic relationship among 13 Chinese and introduced cattle breeds using microsatellite DNA markers[J].BiodiversityScience,2006,14(6):498-507.(in Chinese) [10] CHAKRABORTY R,ZHONG Y,DEANDRADE M,et al.Linkage disequilibria among (CA)n polymorphism in the human dystrophin gene and their implications in carrier detection and prenatal diagnosis in duchenne and becker muscular dystrophies [J].Genomics,1994,21:567-570. [11] BOSTEIN D,WHITE R L,SKOLNICK M,et al.Construction of a genetic linkage map in man using restriction fragment length polymorphisms [J].AmJHumGenet,1980,32:314-331. [12] YANG W J,KANG X L,YANG F Y,et al.Review on the development of genotyping methods for assessing farm animal diversity [J].JAnimSciBiotechnol,2013,4:1-6. [13] GOLDSTEIN D B.An evaluation of genetic distance for use with microsatellite loci [J].Genetics,1995,139:463-471. [14] EBLATE E M,LUGHANO K J,CHENYAMBUGA D,et al.Polymorphic microsatellite markers for genetic studies of African antelope species [J].AfricanJBiotechnol,2011,10(56):11817-11820. [15] 徐云碧.分子數量遺傳學[M].北京:農業出版社,1994. XU Y B.Molecular Quantitative Genetics[M].Beijing:Agricultural Publishing House,1994.(in Chinese) [16] CRONIN M A,MACNEIL M D,VU N, et al.Genetic variation and differentiation of bison (Bison bison) subspecies and cattle (Bostaurus) breeds and subspecies[J].JHeredity,2013,104(4):500-509. [17] KEROS T,JEMERSIC L,PRPIC J, et al. Genetic variability of microsatellites in autochthonous Podolian cattle breeds in Croatia[J].ActaVeterinariaBrno,2013,82(2):135-140. [18] 雷初朝,陳 宏,楊公社,等.中國部分黃牛品種mtDNA遺傳多樣性研究[J].遺傳學報,2004,31(1):43-50. LEI C C,CHEN H,YANG G S,et al.Genetic diversity of Mitochondrial DNA in chinese yellow cattle[J].JournalofGeneticsandGenomics,2004,31(1):43-50.(in Chinese) [19] PHAM L D,DO D N,BINH N T,et al.Assessment of genetic diversity and population structure of Vietnamese indigenous cattle populations by microsatellites[J].LivestSci,2013,155(1):17-22. [20] UTSUNOMIYA Y T,BOMBA L,LUCENTE G,et al.Revisiting AFLP fingerprinting for an unbiased assessment of genetic structure and differentiation of taurine and zebu cattle[J].BMCGenet,2014,15(1):47. [21] ACOSTA C,UFFO O.SANZ A,et al.Genetic diversity and differentiation of five Cuban cattle breeds using 30 microsatellite loci[J].JAnimBreedGenet,2013,130(1):79-86. [22] ARMSTRONG E,IRIARTE A,MARTINEZ A M,et al.Genetic diversity analysis of the Uruguayan Creole cattle breed using microsatellites and mtDNA markers[J].GenetMolRes,2013,12:1119-1131. (編輯 郭云雁) Analyses of Genetic Diversity among Jinnan Cattle and Three other Chinese Indigenous Cattle Breeds WANG Xi1,ZHANG Yuan-qing1*,HE Dong-chang1,ZHANG Xi-zhong1,LI Bo1,WANG Dong-cai1,JIN Guang1,LI Fu-xing2,YANG Xiao-min1,XU Fang1 (1.InstituteofAnimalScienceandVeterinaryMedicine,ShanxiAcademyofAgriculturalScience,Taiyuan030032China;2.YunchengYellowCattleFarm,Yuncheng044500,China) The genetic diversity and genetic structure of Jinnan cattle,Jiaxian Red cattle,Luxi cattle and Qinchuan cattle were detected by microsatellite technology.Allele frequencies and distribution of 16 microsatellite markers were detected in 4 cattle populations.Number of effective alleles of the other 15 loci was from 2 to 8 except HEL9 as a monomorphic loci in all individuals,and the average number of effective alleles was 3.067 6.BM1818 was low genetic polymorphisms (PIC=0.083 0,PIC<0.25),and the others were high genetic polymorphisms (PIC>0.5),with the highestPICvalue at HUAT24 locus (PIC=0.727 5).80 alleles were identified in 4 populations (63 alleles in Jinnan cattle,65 in Jiaxian Red cattle,65 in Luxi cattle and 68 in Qinchuan cattle,respectively).The allele B at IDVGA46 was detected only in Jinnan cattle,and the allele B at TGLA44 was only lack in Jinnan cattle.Mean observed heterozygosity,mean expected heterozygosity and mean heterozygosity were 0.385 2,0.640 3 and 0.578 0,respectively in 4 populations.Jinnan cattle was independently clustered in the NJ tree and the genetic distance (DA) with Luxi cattle,Jiaxian Red cattle and Qinchuan cattle were 0.809 3,0.759 8 and 0.807 1.There were special genetic characterics and sufficient genetic diversity in Jinnan cattle,and it was a relatively closed population in its evolutionary process. microsatellites;genetic diversity;Jinnan cattle 10.11843/j.issn.0366-6964.2015.06.006 2014-07-28 “十二五”國家高技術研究發展計劃(863計劃)子課題(2011AA100307-05);山西省科技攻關項目(20130311024-1;20120311021-1;20130311024-2;20140311018-1);山西省農業科學院育種工程項目(11yzgc017);山西省農業科學院博士基金(YBSJJ1402) 王 曦(1975-),男,山西嵐縣人,副研究員,博士,主要從事牛遺傳育種與功能基因組研究,E-mail:wxphilip@aliyun.com *通信作者:張元慶,副研究員,E-mail:yuanqing_zhang@163.com S813.1;S823 A 0366-6964(2015)06-0911-13

