線粒體DNA D-LOOP區點突變和長期慢性肝炎、肝癌的相關性分析
柳云 劉道潔 喬錄新 陳德喜
[摘要] 目的 通過收集乙型肝炎病毒(HBV)相關的肝癌患者的標本,分析各組織線粒體DNA D-LOOP區堿基的變化,探究HBV相關的肝癌線粒體DNA D-LOOP區的相關性。 方法 選取2008年10月~2019年12月首都醫科大學附屬北京佑安醫院就診的慢性乙肝相關肝細胞癌(HCC)患者44例,其中31例患者的癌組織和對應的癌旁組織及外周血,7例患者的癌旁組織和相對應的外周血,5例患者有癌組織和對應的外周血,11例癌組織和對應的癌旁組織。同時選取了11例來自肝移植患者供體者的肝組織及對應的外周血作為對照。提取標本的線粒體DNA,設計引物和PCR反應獲得線粒體D-LOOP區,對PCR產物進行一代測序。 結果 一代測序數據結果顯示長期慢性HBV肝炎可引起炎癥肝組織和肝癌線粒體DNA D-LOOP區大量單堿基突變,肝癌組織和肝組織之間差異有統計學意義(P < 0.05)。 結論 長期慢性HBV肝炎可引起肝炎組織大量線粒體DNA D-LOOP區堿基突變,肝癌組織不同于正常肝組織的突變模式,對于肝癌的早期診斷具有提示意義。
[關鍵詞] 線粒體DNA D-LOOP區;乙型肝炎病毒;肝細胞癌
[中圖分類號] R512.62? ? ? ? ? [文獻標識碼] A? ? ? ? ? [文章編號] 1673-7210(2019)09(b)-0121-04
Correlation analysis between mitochondrial DNA D-LOOP point mutation and long-term chronic hepatitis and hepatocellular carcinoma
LIU Yun1? ?LIU Daojie2? ?QIAO Luxin1? ?CHEN Dexi1▲
1.Beijing Youan Hospital, Capital Medical University, Beijing Institute of Hepatology, Beijing? ?100069, China; 2.Laboratory Medicine, Haidian Maternal & Child Health Hospital of Beijing City, Beijing? ?100080, China
[Abstract] Objective To analyze the changes in bases of mitochondrial DNA D-LOOP in various tissues by collecting specimens of hepatitis B virus (HBV)-associated liver cancer patients, and to explore the correlation between HBV-associated liver cancer and mitochondrial DNA D-LOOP. Methods Forty-six patients with chronic hepatitis B-related hepatocellular carcinoma (HCC) who were admitted to Beijing Youan Hospital, Capital Medical University from October 2008 to December 2019 were enrolled in the study. Thirty-one cases of cancerous tissues and corresponding adjacent tissues and peripheral blood, and 7 cases of adjacent tissues and corresponding peripheral blood, 5 cases of cancerous tissues and corresponding peripheral blood, 11 cases of cancer tissues and corresponding adjacent tissues. At the same time, 11 liver tissues from donors of liver transplantation patients and corresponding peripheral blood were selected as controls. The mitochondrial DNA of the specimen was extracted, primers and PCR reactions were designed to obtain the mitochondrial D-LOOP region, and the PCR product was sequenced for one generation. Results The results of one-generation sequencing data showed that long-term chronic HBV hepatitis caused a large number of single-base mutations in mitochondrial DNA D-LOOP region of inflammatory liver tissue and liver cancer, and the difference between liver cancer tissues and liver tissues was statistically significant (P < 0.05). Conclusion Long-term chronic HBV hepatitis can cause a large number of mitochondrial DNA D-LOOP region mutations in hepatitis tissues. Hepatoma tissue is different from normal liver tissue mutation mode, which has implications for the early diagnosis of liver cancer.
[Key words] Mitochondrial DNA D-LOOP region; Hepatitis B virus; Hepatocellular carcinoma
肝癌是世界上最常見的惡性腫瘤之一,其發病率位居男性第五位,女性位居第九位,死亡率在人群中居第二位[1]。肝細胞癌(HCC)是肝癌中最常見的組織學類型[2]。HBV感染和慢性肝病是HCC發生的主要危險因素[3]。
肝臟的功能依賴于線粒體的代謝穩態[4-6]。線粒體DNA(mtDNA)是一個16.6 kb的雙鏈環狀DNA[7-8]。由于線粒體內富含ROS且mtDNA缺乏組蛋白的保護,缺少有效、完整的修復系統[11]。mtDNA序列微小的變化可以導致功能上的嚴重損害[9]。mtDNA突變的累積有助于腫瘤的起始和進展[10-11]。mtDNA“D環”的非編碼區,控制mtDNA的復制和轉錄[12]。線粒體的轉錄和翻譯功能的改變與肝癌相關[13],在人類癌癥中發現的大多mtDNA突變位于D-環區域,且報道與肝癌發生密切相關[14-15]。
線粒體DNA突變與肝癌的關系,已有很多橫斷面的研究[16-20],但是長期HBV慢性炎癥引起的細胞ROS升高如何引起線粒體DNA的損傷累集,至今仍鮮見報道。本研究中以患者外周血PBMC的mtDNA序列為線粒體遺傳對照,分析長期慢性肝炎及相關肝癌mtDNA突變積累特征,并以肝移植供體的肝臟組織及PBMC的線粒體DNA作為對照進行對比分析,闡明HBV慢性肝炎引起肝組織及肝癌mtDNA D-LOOP的突變深度和特征,對了解慢性肝炎進展程度和肝癌發生有一定提示意義。
1 資料與方法
1.1 一般資料
收集首都醫科大學附屬北京佑安醫院(以下簡稱“我院”)就診的初診HCC患者43例,其中31例患者同時具有癌組織和對應的癌旁組織及外周血,7例患者有癌旁組織和相對應的外周血,5例患者有癌組織和對應的外周血,11例對照來自肝移植患者供體者的肝組織及對應的外周血。本研究標本為國家“十二五”項目收集的標本,項目所需人源組織標本已獲我院醫學倫理學委員會批準。肝穿組織獲取后立即置于液氮中儲存。NAA提取使用凱杰DNA提取試劑盒,根據制造商的說明書提取總基因組DNA。提取的DNA存儲在-20℃備用。
1.2 方法
擴增所用引物見表1,使用Mt-D-LOOP S1、AS1擴增產物為1332 bp mtDNA D-LOOP片段。聚合酶鏈式反應(PCR)在50 μL的體系中進行,包括mix 25 μL,H2O 18 μL,前后引物各1 μL,mtDNA 1 μL。整個實驗過程于冰上操作。PCR反應條件:94℃ 2 min,35個循環;94℃ 15 s,52℃ 30 s,72℃ 1 min 30 s,最終72℃下延伸5 min。通過1% TAE瓊脂糖凝膠電泳分析PCR產物。PCR產物進行一代測序,測序引物為Mt-D-LOOP S1、AS1、S2、AS2、S3、AS3,由北京博邁德公司進行PCR產物測序。
表1? ?擴增和測序引物列表
1.3 統計學方法
mtDNA標準序列為mtDNA修訂劍橋序列,將患者中序列的變異視為突變,供體的變異視為多態性。序列分析使用SanpGene。進化樹作圖使用MEGA6軟件,使用Prism統計學軟件進行數據統計分析,使用Two-way ANOVA分析組間差異,以P < 0.05為差異有統計學意義。
2 結果
2.1 PCR實驗獲得穩定特異的目的條帶
PCR實驗得到了穩定特異行mtDNA D-LOOP區片段,經1%的TAE瓊脂糖膠進行電泳分析。見圖1。
2.2 所有樣本的進化樹分析結果
進化樹分析,Bootstrap的值>70%,來自同一患者的標本分布于同一支,提示PCR產物無交叉污染,見圖2。與外周PBMC比較,同一患者的肝癌和肝組織距離標準序列更遠,而10例正常肝臟與PBMC的距離非常接近,提示長期慢性肝病可以影響肝組織的mtDNA D-LOOP的突變水平。完整的進化樹圖見圖2。
2.3 來自肝移植供體肝臟和對應外周血之間mtDNA D-LOOP測序比較
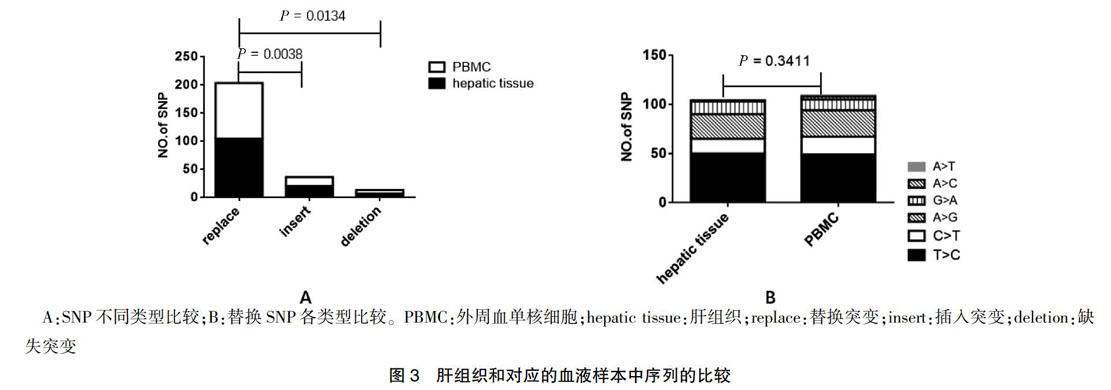
11個供體者的肝臟組織和對應的外周血中共觀察到66個SNP位點,262個SNP。替換突變數量多于其他兩種類型,差異有統計學意義(P = 0.0038、0.0134);供體的肝臟和對應的外周血替換SNP各類型比較差異無統計學意義(P = 0.3411)。見圖3。
2.4 對31對完全配對HCC患者標本堿基序列比較
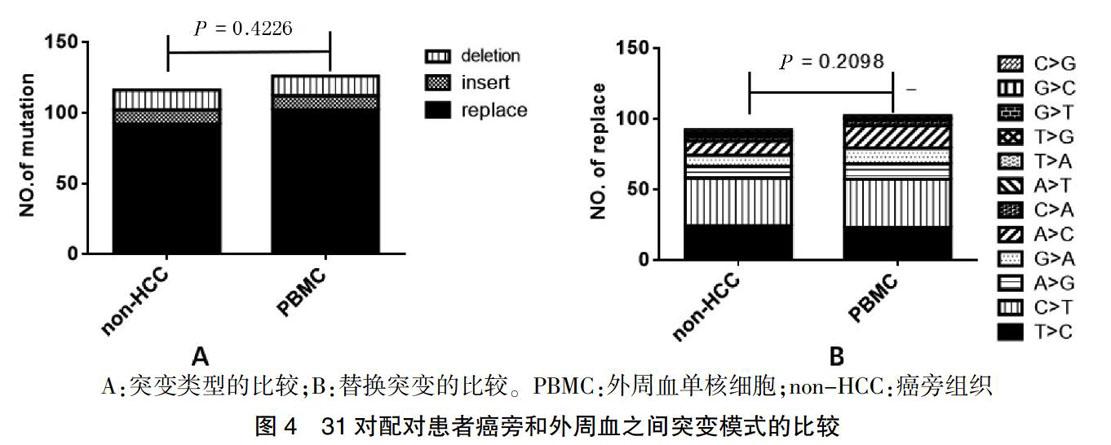
31個配對患者的HCC組織、癌旁組織和外周血中的突變類型和替換突變之間的差異分析,發現在同一患者中,不管是突變的類型還是替換突變的各類型之間,癌旁組織和對應的外周血之間差異無統計學意義(P = 0.4226、0.2098)。見圖4。
2.5 所有患者標本堿基序列比較情況
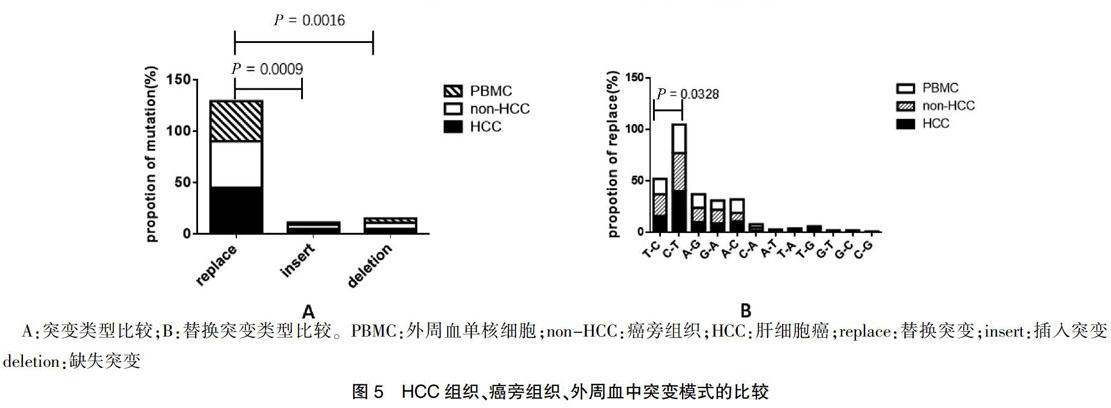
本研究分析了患者癌組織、非癌組織、外周血液組織,3種組織中替代突變明顯高于插入突變和缺失突變,差異有統計學意義(P = 0.0009、0.0016),見圖5A,且C 3 討論 線粒體是體內唯一具有獨立DNA的供能細胞器,與肝癌的發生發展具有密切關系。細胞mtDNA可能導致缺陷性線粒體的積累。目前研究表明,mtDNA可用于重癥肝炎患者肝細胞的損傷評估。有研究證實肝癌患者血漿mtDNA表達量亦發生改變。由于mtDNA具有長度短、分子結構簡單和拷貝數高等特點,使得監控早期腫瘤患者mtDNA的異常變化成為可能。相鄰的癌旁組織的mtDNA并不能認為是正常的mtDNA。與以往的研究比較,本研究首次系統全面地收集組織的類型,系統多層次地比較不同組織類型的突變模式的差異,并且首次收集并使用了供體的肝臟和對應的外周血,為正常肝臟mtD-LOOP區,提供了寶貴且真實的線粒體正常肝臟序列。 本研究進一步證明了mtDNA D-LOOP區突變在HBV相關的慢性肝炎和肝硬化的過程,觀察到癌組織和相鄰的癌旁組織之間存在突變模式的差異,而癌旁組織和外周血之間沒有顯著的差異。本研究提示HCC和炎性肝組織中mtDNA的不同誘變機制以及肝癌發生過程中可能存在進化選擇,在癌癥的進化過程中存在著對于這些突變的陽性選擇,突變模式的不同對肝癌的診斷有一定提示意義。 [參考文獻] [1]? Torre LA,Bray F,Siegel RL,et al. Global cancer statistics,2012 [J]. CA Cancer J Clin,2015,65(2):87-108. [2]? Perz JF,Armstrong GL,Farrington,LA,et al. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide [J]. J Hepatol,2006,45(4):529-538. [3]? Yuan K,Lei Y,Chen HN,et al. HBV-induced ROS accumulation promotes hepatocarcinogenesis through Snail-mediated epigenetic silencing of SOCS3 [J]. Cell Death Differ,2016,23(4):616-627. [4]? Zhao Y,Liu S,Zhou L,et al. Aberrant shuttling of long noncoding RNAs during the mitochondria-nuclear crosstalk in hepatocellular carcinoma cells [J]. Am J Cancer Res,2019,9(5):999-1008. [5]? Qiao L,Nie Z,Li Q,et al. Mitochondrial DNA depletion,mitochondrial mutations and high TFAM expression in hepatocellular carcinoma [J]. Oncotarget,2017,8(48):84 373-84 383. [6]? Hikita H,Kodama T,Tanaka S,et al. Activation of the Mitochondrial Apoptotic Pathway Produces Reactive Oxygen Species and Oxidative Damage in Hepatocytes That Contribute to Liver Tumorigenesis [J]. Cancer Prev Res (Phila),2015,8(8):693-701. [7]? Attardi G,Schatz G. Biogenesis of mitochondria [J]. Annu Rev Cell Biol,1988,4:289-333. [8]? Anderson S,Bankier AT,Barrell BG,et al. Sequence and organization of the human mitochondrial genome [J]. Nature,1981,290(5806):457-465. [9]? Nishikawa M,Nishiguchi S,Kioka K,et al. Interferon reduces somatic mutation of mitochondrial DNA in liver tissues from chronic viral hepatitis patients [J]. J Viral Hepat,2005,12(5):494-498. [10]? Li X,Guo X,Li D,et al. Multi-regional sequencing reveals intratumor heterogeneity and positive selection of somatic mtDNA mutations in hepatocellular carcinoma and colorectal cancer [J]. Int J Cancer,2018,143(5):1143-1152. [11]? Yu C,Wang X,Huang L,et al. Deciphering the Spectrum of Mitochondrial DNA Mutations in Hepatocellular Carcinoma Using High-Throughput Sequencing [J]. Gene Expr,2018,18(2):125-134. [12]? Attardi G,Schatz G. Biogenesis of mitochondria [J]. Annu Rev Cell Biol,1988,4:289-333. [13]? Baechler SA,Factor VM,Dalla Rosa I,et al. The mitochondrial type IB topoisomerase drives mitochondrial translation and carcinogenesis [J]. Nat Commun,2019,10(1):83. [14]? Li S,Wan P,Peng T,et al. Associations between sequence variations in the mitochondrial DNA D-loop region and outcome of hepatocellular carcinoma [J]. Oncol Lett,2016,11(6):3723-3728. [15]? Yin C,Cao HY,Chen YB,et al. NGS-based profiling reveals a critical contributing role of somatic D-loop mtDNA mutations in HBV-related hepatocarcinogenesis [J]. Ann Oncol,2019,30(6):953-962. [16]? Chen T,Xun Z,Lin J,et al. Association between mitochondrial DNA content and baseline serum levels of HBsAg in chronic hepatitis B infection [J]. J Med Virol,2017,89(11):1958-1962. [17]? Li L,Hann HW,Wan S,et al. Cell-free circulating mitochondrial DNA content and risk of hepatocellular carcinoma in patients with chronic HBV infection [J]. Sci Rep,2016,6(1):23 992. [18]? Campo DS,Nayak V,Srinivasamoorthy G,et al. Entropy of mitochondrial DNA circulating in blood is associated with hepatocellular carcinoma [J]. BMC Med Genomics,2019,12(4):74. [19]? Guo ZS,Jin CL,Yao ZJ,et al. Analysis of the Mitochondrial 4977 Bp Deletion in Patients with Hepatocellular Carcinoma [J]. Balkan J Med Genet,2017,20(1):81-86. [20]? Gao Y,Nie HJ,Yang D,et al. Changes of the mitochondrial DNA copy number and the antioxidant system in the PBMC of hepatocellular carcinoma [J]. Zhongguo Ying Yong Sheng Li Xue Za Zhi,2016,32(1):1-5. (收稿日期:2018-06-05? 本文編輯:任? ?念)

